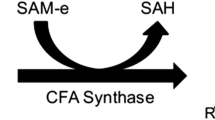
Abstract
Past research has sought to improve the production of cyclopropane fatty acids by the oleaginous yeast Yarrowia lipolytica by heterologously expressing the E. coli fatty acid synthase gene and improving cultivation processes. Cyclopropane fatty acids display properties that hold promise for biofuel applications. The E. coli fatty acid synthase gene was introduced into several genetic backgrounds of the yeast Y. lipolytica to optimize lipid synthesis; the mean cyclopropane fatty acid productivity was 43 mg L−1 h−1 on glucose, and the production rate reached its maximum (3.06 g L−1) after 72 h of cultivation in a bioreactor. The best strain (JMY6851) overexpressed simultaneously the E. coli cyclopropane fatty acid synthase gene under a hybrid promoter (hp8d) and Y. lipolytica LRO1 gene. In fed-batch process using crude glycerol as carbon source, JMY6851 strain displayed high lipid accumulation (78% of dry cell weight) and high biomass production (56 g L−1). After 165 h of cultivation, cyclopropane fatty acids represented 22% of the lipids produced; cyclopropane fatty acid productivity (103.3 mg L−1 h−1) was maximal at 72.5 h of cultivation.




Similar content being viewed by others
References
Beopoulos A, Cescut J, Haddouche R et al (2009) Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res 48:375–387. https://doi.org/10.1016/j.plipres.2009.08.005
Beopoulos A, Nicaud JM (2012) Yeast : A new oil producer ? OCL-Ol Corps Gras Lipides 19:22–28. https://doi.org/10.1684/ocl.2012.0426
Ratledge C, Wynn J (2002) The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol 50:1–51
Groenewald M, Boekhout T, Neuvéglise C et al (2014) Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit Rev Microbiol 40:187–206
Rywińska A, Juszczyk P, Wojtatowicz M et al (2013) Glycerol as a promising substrate for Yarrowia lipolytica biotechnological applications. Biomass Bioenergy 48:148–166. https://doi.org/10.1016/j.biombioe.2012.11.021
Larroude M, Celinska E, Back A et al (2018) A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol Bioeng. https://doi.org/10.1002/bit.26473
Ledesma-Amaro R, Nicaud JM (2016) Metabolic engineering for expanding the substrate range of Yarrowia lipolytica. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2016.04.010
Lazar Z, Neuvéglise C, Rossignol T et al (2017) Characterization of hexose transporters in Yarrowia lipolytica reveals new groups of sugar porters involved in yeast growth. Fungal Genet Biol. https://doi.org/10.1016/j.fgb.2017.01.001
Beopoulos A, Nicaud JM, Gaillardin C (2011) An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl Microbiol Biotechnol 90:1193–1206. https://doi.org/10.1007/s00253-011-3212-8
Dulermo T, Nicaud JM (2011) Involvement of the G3P shuttle and Β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab Eng 13:482–491. https://doi.org/10.1016/j.ymben.2011.05.002
Albertyn J, Hohmann S, Thevelein JM, Prior BA (1994) GPD1, which encodes Glycerol-3-Phosphate Dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol 14:4135–4144
Beopoulos A, Mrozova Z, Thevenieau F et al (2008) Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Env Microbiol 74:7779–7789. https://doi.org/10.1128/AEM.01412-08
Morash SC, Mcmaster CR, Hjelmstad RH et al (1994) Studies employing Saccharomyces cerevisiae CPT1 and EPT1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis. J Biol Chem 269:28769–28776
Ledesma Amaro R, Nicaud JM (2016) Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog Lipid Res 61:40–50. https://doi.org/10.1016/j.plipres.2015.12.001
Wang HJ, Le Dall MT, Waché Y et al (1999) Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol 181:5140–5148
Dulermo T, Tréton B, Beopoulos A et al (2013) Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: New insights into the role of intracellular lipases and lipid body organisation. Biochim Biophys Acta 1831:1486–1495. https://doi.org/10.1016/j.bbalip.2013.07.001
Friedlander J, Tsakraklides V, Kamineni A et al (2016) Engineering of a high lipid producing Yarrowia lipolytica strain. Biotechnol Biofuels 9:1–12. https://doi.org/10.1186/s13068-016-0492-3
Gajdos P, Nicaud JM, Rossignol T, Certik M (2015) Single cell oil production on molasses by Yarrowia lipolytica strains overexpressing DGA2 in multicopy. Appl Microbiol Biotechnol 99:8065–8074. https://doi.org/10.1007/s00253-015-6733-8
Beopoulos A, Haddouche R, Kabran P et al (2012) Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl Microbiol Biotechnol 93:1523–1537. https://doi.org/10.1007/s00253-011-3506-x
Beopoulos A, Verbeke J, Bordes F et al (2014) Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 98:251–262. https://doi.org/10.1007/s00253-013-5295-x
Ledesma-Amaro R, Dulermo R, Niehus X, Nicaud J-M (2016) Combining metabolic engineering and process optimization to improve production and secretion of fatty acids. Metabol Eng. https://doi.org/10.1016/j.ymben.2016.06.004
Svensson L, Hansson U, Gronowitz S, Klingstedt T (1997) The relationship between the structure of monoalkyl branched saturated triacylglycerols and some physical properties. Lipids 32:661–666. https://doi.org/10.1007/s11745-997-0084-2
Cronan JE, Reed R, Taylor FR, Jacksont MB (1979) Properties and biosynthesis of cyclopropane fatty acids in Escherichia coli. J Bacteriol 138:118–121
Duhot P, Gontier E, Thomas D, et al. (1999) Procedé de production d’acides gras ramifiés au moyen de plantes genétiquement modifiées. Patent PCT/FR 9802116. Europe, Canada, Etats Unis, Japon, WOFR9802116
Cronan JE, Nunn WD, Batchlor JG (1974) Studies on the biosynthesis of cyclopropane fatty acids in Escherichia coli. Biochim biophys acta 348:63–75
Grogan DW, Cronan JE (1997) Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev MMBR 61:429–441
Montanari C, Sado Kamdem SL, Serrazanetti DI et al (2010) Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol 27:493–502. https://doi.org/10.1016/j.fm.2009.12.003
Czerwiec Q, Idrissi Taghki A, Imatoukene N et al (2019) Optimisation of cyclopropane fatty acids production in Yarrowia lipolytica. Yeast 36:143–151. https://doi.org/10.1002/yea.3379
Grogan DW, Cronan JE (1984) Cloning and manipulation of the Escherichia coli cyclopropane fatty acid synthase gene : physiological aspects of enzyme overproduction. J Bacteriol 158:286–295
Markham KA, Alper HS (2018) Engineering Yarrowia lipolytica for the production of cyclopropanated fatty acids. J Ind Microbiol Biotechnol. https://doi.org/10.1007/s10295-018-2067-8
Bao X, Katz S, Pollard M, Ohlrogge J (2002) Carbocyclic fatty acids in plants: biochemical and molecular genetic characterization of cyclopropane fatty acid synthesis of Sterculia foetida. Proc Natl Acad Sci USA 99:7172–7177. https://doi.org/10.1073/pnas.092152999
Bao X, Thelen JJ, Bonaventure G, Ohlrogge JB (2003) Characterization of cyclopropane fatty-acid synthase from Sterculia foetida. J Biol Chem 278:12846–12853. https://doi.org/10.1074/jbc.M212464200
Zhao Y, Hindorff L, Chuang A et al (2003) Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 69:2831–2841. https://doi.org/10.1128/AEM.69.5.2831
Yu X-H, Rawat R, Shanklin J (2011) Characterization and analysis of the cotton cyclopropane fatty acid synthase family and their contribution to cyclopropane fatty acid synthesis. Plant Biol 11:97. https://doi.org/10.1186/1471-2229-11-97
Yu X, Prakash RR, Sweet M, Shanklin J (2014) Coexpressing Escherichia coli cyclopropane synthase with Sterculia foetida lysophosphatidic acid acyltransferase enhances cyclopropane fatty acid accumulation. Plant Physiol 164:455–465. https://doi.org/10.1104/pp.113.230953
Machida S, Shiraiwa Y, Suzuki I (2016) Construction of a cyanobacterium synthesizing cyclopropane fatty acids. Biochim Biophys Acta 1861:980–987. https://doi.org/10.1016/j.bbalip.2016.05.012
Muller Sven ST (1998) Comparison of expression systems in the yeasts Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast 14:1267–1283
Madzak C, Blanchin-Roland S, Cordero Otero RR, Gaillardin C (1999) Functional analysis of upstream regulating regions from the Yarrowia lipolytica XPR2 promoter. Microbiology 145:75–87
Dulermo R, Brunel F, Dulermo T et al (2017) Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microb Cell Fact 16:31. https://doi.org/10.1186/s12934-017-0647-3
Barth G, Gaillardin C (1996) Yarrowia lipolytica. In: Wolf K (ed) Non conventional yeasts in biotechnology. Springer, Berlin, pp 314–388
Lazar Z, Dulermo T, Neuvéglise C et al (2014) Hexokinase a limiting factor in lipid production from fructose in Yarrowia lipolytica. Metab Eng 26:89–99. https://doi.org/10.1016/j.ymben.2014.09.008
Sagnak R, Cochot S, Molina-Jouve C et al (2018) Modulation of the glycerol phosphate availability led to concomitant reduction in the citric acid excretion and increase in lipid content and yield in Yarrowia lipolytica. J Biotechnol 265:40–45. https://doi.org/10.1016/j.jbiotec.2017.11.001
Dulermo R, Gamboa-Meléndez H, Dulermo T et al (2014) The fatty acid transport protein Fat1p is involved in the export of fatty acids from lipid bodies in Yarrowia lipolytica. Yeast Res 14:883–896. https://doi.org/10.1111/1567-1364.12177
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Back A, Rossignol T, Krier F et al (2016) High-throughput fermentation screening for the yeast Yarrowia lipolytica with real-time monitoring of biomass and lipid production. Microb Cell Fact 15:147. https://doi.org/10.1186/s12934-016-0546-z
Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509
Browse J, Mccourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152:141–145
Park YK, Dulermo T, Amaro RL, Nicaud JM (2018) Biotechnology for biofuels optimization of odd chain fatty acid production by Yarrowia lipolytica. Biotechnol Biofuels 11:1–12. https://doi.org/10.1186/s13068-018-1154-4
Merlier F, Imatoukene N, Octave S et al (2018) Spectrometry method for separation and characterization of 3-hydroxymethyl pyridine ester of fatty acids at low levels. J Chromatogr A 1575:72–79. https://doi.org/10.1016/j.chroma.2018.09.010
Sumner J, Noback C (1924) The estimation of sugar in diabetic urine, using dinitrosalicylic acid. J Biol Chem 62:290–297
Xu J, Zhao X, Wang W et al (2012) Microbial conversion of biodiesel byproduct glycerol to triacylglycerols by oleaginous yeast Rhodosporidium toruloides and the individual effect of some impurities on lipid production. Biochem Eng J 65:30–36. https://doi.org/10.1016/j.bej.2012.04.003
Chatzifragkou A, Papanikolaou S (2012) Effect of impurities in biodiesel-derived waste glycerol on the performance and feasibility of biotechnological processes. Appl Microbiol Biotechnol 95:13–27. https://doi.org/10.1007/s00253-012-4111-3
Samul D, Leja K, Grajek W (2014) Impurities of crude glycerol and their effect on metabolite production. Ann Microbiol 64:891–898. https://doi.org/10.1007/s13213-013-0767-x
Hardman D, McFalls D, Fakas S (2017) Characterization of phosphatidic acid phosphatase activity in the oleaginous yeast Yarrowia lipolytica and its role in lipid biosynthesis. Yeast 34:83–91. https://doi.org/10.1002/yea.3216
Acknowledgements
This work was performed in collaboration with SAS PIVERT (www.institut-pivert.com), which is an institute for energy transition (Institut pour la Transition Energétique [ITE] PIVERT)). SAS PIVERT received funding from the French government’s Investments for the Future programme (Investissements d’Avenir). This study was also supported by Investments for the Future funding (ANR-001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Imatoukene, N., Back, A., Nonus, M. et al. Fermentation process for producing CFAs using Yarrowia lipolytica. J Ind Microbiol Biotechnol 47, 403–412 (2020). https://doi.org/10.1007/s10295-020-02276-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-020-02276-6