Abstract
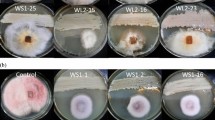
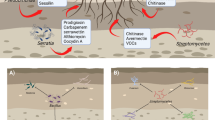
The present study investigated the antagonistic and plant growth promoting (PGP) potential of actinobacteria TT3 isolated from tea rhizosphere soil of Tocklai tea germplasm preservation plot, Jorhat, Assam, India. It is a Gram-positive, filamentous with flexible spore chains actinobacteria. The 16S rRNA gene sequencing and phylogenetic analysis indicated that TT3 is closely related to genus Streptomyces for which it was referred to as Streptomyces sp. TT3. It showed very promising PGP traits such as phosphate solubilization, production of indole acetic acid (IAA), siderophore, and ammonia. Evaluation of ethyl acetate extract of TT3 exhibited broad spectrum antagonistic activity against various fungal pathogens. This antagonistic Streptomyces sp. TT3 showed positive for polyketide synthase type II (PKS-II) gene, which was predicted to be involved in the production of actinorhodin as a secondary metabolite pathway product using DoBISCUIT database. Further, the crude ethyl acetate extract of TT3 was analyzed by using GC–MS and revealed the presence of significant chemical constituents responsible for antimicrobial activity. Thus, the present study suggests that actinobacteria isolated from the rhizosphere soil may be explored for the production of bioactive compounds and use as a potential candidate for tea and other agricultural application.

Similar content being viewed by others
References
Calderón K, Spor A, Breuil MC, Bru D, Bizouard F, Violle C, Barnard RL, Philippot L (2016) Effectiveness of ecological rescue for altered soil microbial communities and functions. ISME 16:1751–7362
Compant S, Clément C, Sessitsch A (2010) Plant growth promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678
Hamedi J, Mohammadipanah F (2015) Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J Ind Microbiol Biotechnol 42:157–171
Shivlata L, Satyanarayana T (2017) Actinobacteria in agricultural and environmental sustainability. In: Singh JS, Seneviratne G (eds) Agro environmental sustainability. Springer, Berlin, Heidelberg, pp 173–218
Palaniyandi SA, Yang SH, Zhang L, Suh JW (2013) Effects of actinobacteria on plant disease suppression and growth promotion. Appl Microbiol Biotechnol 97:9621–9636
Sathya A, Vijayabharathi R, Gopalakrishnan S (2017) Plant growth-promoting actinobacteria: a new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 7:102
Majumder AB, Bera B, Rajan A (2010) Tea statistics: global scenario. Inc J Tea Sci 8:121–124
Barthakur BK (2011) Recent approach of Tocklai to plant protection in tea in Northeast India. Sci Cult 77:381–384
Chakraborty U, Chakraborty BN, Basnet M (2006) Plant growth promotion and induction of resistance in Camellia sinensis by Bacillus megaterium. J Basic Microbiol 46:186–195
Chakraborty U, Chakraborty BN, Chakraborty AP, Sunar K, Dey PL (2013) Plant growth promoting rhizobacteria mediated improvement of health status of tea plants. Indian J Biotechnol 12:20–31
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Kimura MA (1980) Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fiske CH, Subbarow Y (1925) A colorimetric determination of phosphorus. J Biol Chem 66:375–400
Ryu RJ, Patten CL (2008) Aromatic amino acid-dependent expression of indole-3-pyruvate decarboxylase is regulated by TyrR in Enterobacter cloacae UW5. J Bacteriol 190:7200–7208
Gordon SA, Weber RP (1951) Colorimetric estimation of indole-acetic acid. Plant Physiol 26:192–195
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Patel AK, Deshattiwar MK, Chaudhari BL, Chincholkar SB (2009) Production, purification and chemical characterization of the catecholate siderophore from potent probiotic strains of Bacillus spp. Bioresour Technol 100:368–373
Goswami D, Dhandhukia P, Patel P, Thakker JN (2014) Screening of PGPR from saline desert of Kutch: Growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol Res 169:66–75
El-Sayed WS, Akhkha A, El-Naggar MY, Elbadry M (2014) In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front Microbiol 5:651. https://doi.org/10.3389/fmicb.2014.00651
Nweze EI, Mukherjee PK, Ghannoum MA (2010) Agar-based disk diffusion assay for susceptibility testing of dermatophytes. J Clin Microbiol 48:3750–3752
Ayuso-Sacido A, Genilloud O (2005) New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol 49:10–24
Ichikawa N, Sasagawa M, Yamamoto M, Komaki H, Yoshida Y, Yamazaki S, Fujita N (2013) DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res 41:408–414
Ser H, Palanisamy UD, Yin WF, AbdMalek SN, Chan KG, Goh BH, Lee LH (2015) Presence of antioxidative agent, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro in newly isolated Streptomyces mangrovisoli sp. Nov. Front Microbiol 6:854. https://doi.org/10.3389/fmicb.2015.00854
Benizri E, Baudoin E, Guckert A (2001) Root colonization by inoculated plant growth promoting rhizobacteria. Biocontrol Sci Technol 11:557–574
Mitra A, Santra SC, Mukherjee J (2008) Distribution of actinomycetes, their antagonistic behaviour and the physico chemical characteristics of the world’s largest tidal mangrove forest. Appl Microbiol Biotechnol 80:685–695
Yuan WM, Crawford DL (1995) Characterization of Streptomyces lydicus WYEC108 as a potential biocontrol agent against fungal root and seed rots. Appl Environ Microbiol 61:3119–3128
Wang G, Raaijmakers JM (2004) Antibiotics production by bacterial agents and its role in biological control. Chin J Appl Ecol 15:1100–1104
Sansinenea E, Ortiz A (2011) Secondary metabolites of soil Bacillus spp. Biotechnol Lett 33:1523–1538
Leifert C, Li H, Chidburee S, Hampson S, Workman S, Sigee D, Epton HA, Harbour A (1995) Antibiotic production and biocontrol activity by Bacillus subtilis CL27 and Bacillus pumilus CL45. J Appl Bacteriol 78:97–108
Selvin J, Sathiyanarayanan G, Lipton AN, Al-Dhabi NA, Valan Arasu M, Kiran GS (2016) Ketide synthase (KS) domain prediction and analysis of iterative type II PKS gene in marine sponge-associated actinobacteria producing biosurfactants and antimicrobial agents. Front Microbiol 7:63. https://doi.org/10.3389/fmicb.2016.00063
Metsa-Ketela M, Salo V, Halo L, Hautala A, Hakala J, Mantsla P, Ylihonko K (1999) An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol Lett 180:1–6
Fernández-Moreno MA, Martinez E, Boto L, Hopwood DA, Malpartida F (1992) Nucleotide sequence and deduced functions of a set of co-transcribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem 267:19278–19290
Bibb MJ, Biró S, Motamedi H, Collins JF, Hutchinson CR (1989) Analysis of the nucleotide sequence of the Streptomyces glaucescens tcmI genes provides key information about the enzymology of polyketide antibiotic biosynthesis. EMBO J 8:2727–2736
Summers RG, Wendt-Pienkowski E, Motamedi H, Hutchinson CR (1992) Nucleotide sequence of the tcmII-tcmIV region of the tetracenomycin C biosynthetic gene cluster of Streptomyces glaucescens and evidence that the tcmN gene encodes a multifunctional cyclase-dehydratase-O-methyl transferase. J Bacteriol 174:1810–1820
Wawrik B, Kerkhof L, Zylstra GJ, Kukor JJ (2005) Identification of unique type II polyketide synthase genes in soil. Appl Environ Microbiol 71:2232–2238
Cho JY, Kwon HC, Williams PG, Kauffman CA, Jensen PR, Fenical W (2006) Actinofuranones A and B, polyketides from a marine-derived bacterium related to the genus Streptomyces (Actinomycetales). J Nat Prod 69:425–428
Fernando WGD, Ramarathnama R, Krishnamoorthyb AS, Savchuk SC (2005) Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol Biochem 37:955–964
Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186:1084–1096
Pollak FC, Berger RG (1996) Geosmin and related volatiles in bioreactor-cultured Streptomyces citreus CBS 109.60. Appl Environ Microbiol 62:1295–1299
Jog R, Pandya M, Nareshkumar G, Rajkumar S (2014) Mechanism of phosphate solubilisation and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiol 160:778–788
Sengupta S, Pramanik A, Ghosh A, Bhattacharyya M (2015) Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol 15:170. https://doi.org/10.1186/s12866-015-0495-4
Ren J, Wang J, Karthikeyan S, Liu H, Cai J (2019) Natural anti-phytopathogenic fungi compound phenol, 2, 4-bis (1, 1-dimethylethyl) from Pseudomonas fluorescens TL-1. IJBB 56:162–168
Padmavathi AR, Abinaya B, Pandian SK (2014) Phenol, 2,4-bis(1,1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling 30:1111–1122
Kumari N, Menghani E, Mithal R (2019) GCMS analysis of compounds extracted from actinomycetes AIA6 isolates and study of its antimicrobial efficacy. IICT 26:362–370
Pooja S, Aditi T, Naine SJ, Devi CS (2017) Bioactive compounds from marine Streptomyces sp. VITPSA as therapeutics. Front Biol 12:280–289
Kiran GS, Priyadharsini S, Sajayan A, Ravindrana A, Selvin J (2018) An antibiotic agent pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro isolated from a marine bacteria Bacillus tequilensis MSI45 effectively controls multi-drug resistant Staphylococcus aureus. RSC Adv 8:17837–17846
Berdy J (2012) Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot 65:385–395
Acknowledgements
This work was supported by the Department of Biotechnology (DBT), Govt. of India under RGYI scheme (Grant No. BT/PR6011/GBD/27/379/2012). The authors wish to thank Director, IASST, Guwahati, Assam, India, for providing facilities for this work.
Funding
This work was supported by Department of Biotechnology (DBT), Govt. of India under RGYI scheme (Grant No. BT/PR6011/GBD/27/379/2012).
Author information
Authors and Affiliations
Contributions
DT conceived and supervised the research work. DT and JD contributed in the experimental design and interpretation of the data. JD performed the laboratory experiments and data analysis. JD and DT wrote the paper and participated in the revisions of it. Both the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dutta, J., Thakur, D. Evaluation of Antagonistic and Plant Growth Promoting Potential of Streptomyces sp. TT3 Isolated from Tea (Camellia sinensis) Rhizosphere Soil. Curr Microbiol 77, 1829–1838 (2020). https://doi.org/10.1007/s00284-020-02002-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02002-6