Abstract
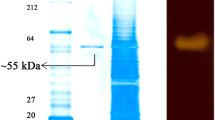
Gracilibacillus dipsosauri is a moderately-halophilic Gram-positive bacterium which forms an extracellular α-amylase that is induced by starch, repressed by d-glucose, and active in 2.0 M KCl. Previous studies showed that while enzyme activity could be measured with the synthetic substrate 2-chloro-4-nitrophenyl-α-d-maltotrioside (CNPG3), other assays were inconsistent and the protein showed aberrant mobility during nondenaturing gel electrophoresis. To clarify the properties of this enzyme, the genome of G. dipsosauri was sequenced and was found to be 4.19 Mb in size with an overall G+C content of 36.9%. A gene encoding an α-amylase composed of 691 amino acids was identified. The protein was a member of the glycosyl hydrolase 13 family, which had a molecular mass of 77,396 daltons and a pI of 4.39 due to an unusually large number of aspartate and glutamate residues (95/691 or 13.7%). BLAST analysis of the amino acid sequence revealed significant matches to other proteins with cyclodextrin glycosyltransferase activity. Partial purification of the protein from G. dipsosauri showed that fractions catalyzing the hydrolysis of CNPG3 and p-nitrophenyl-d-maltoheptoside also catalyzed the formation of β-cyclodextrin but not α-cyclodextrin or γ-cyclodextrin. Formation of β-cyclodextrin was not stimulated by high salt concentrations but did occur with rice, potato, wheat, and corn starches and amylopectin. These studies explain the unusual features of the α-amylase from G. dipsosauri and indicate it should be classified as EC 2.4.1.19. The availability of the complete genomic sequence of G. dipsosauri will provide the basis for studies on other enzymes from this halophile which may be useful for biotechnology.



Similar content being viewed by others
References
Ahmed I, Yokota A, Fugiwara T (2007) Gracilibacillus boraciitolerans sp. nov., a highly boron tolerant and moderately halotolerant bacterium isolated from soil. Int J Sys Evol Microbiol 57:796–802
Basappa C, Rao P, Rao DN, Divakar S (1998) A modified colorimetric method for the estimation of β-cyclodextrin using phenolphthalein. Int J Food Sci Technol 33:517–520
Bernfeld P (1955) Amylases, α and β. Methods Enzymol 1:149–158
Canganella F, Wiegel J (2011) Extremophiles: from abyssal to terrestrial ecosystems and possibly beyond. Naturwissenschaften 98:253–279
Carrasco IJ, Márquez MC, Yanfen X, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A (2006) Gracilibacillus orientalis sp. nov., a novel moderately halophilic bacterium from a salt lake in Inner Mongolia. China. Int J Syst Evol Microbiol 56:599–604
Chamroensaksri N, Tanasupawat S, Akaracharanya A, Visessanguan W, Kudo T, Ito T (2010) Gracilibacillus thailandensis sp. nov., from fermented fish (pla-ra). Int J Syst Evol Microbiol 60:944–948
Chen YG, Cui XL, Zhang YQ, Li WJ, Wang YX, Xu LH, Peng Q, Wen ML, Jiang CL (2008a) Gracilibacillus quinghaiensis sp. nov., isolated from salt-lake sediment in the Qaidam Basin, north-west China. Sys Appl Microbiol 31:181–189
Chen YG, Cui XL, Zhang YQ, Li WJ, Wang YX, Xu LH, Peng Q, Wen ML, Jiang CL (2008b) Gracilibacillus halophilus sp. nov., a moderately halophilic bacterium isolated from saline soil. Int J Sys Evol Microbiol 58:2403–2408
Dalmaso GZL, Ferreira D, Vermelho AB (2015) Marine extremophiles: a source of hydrolases for biotechnological applications. Mar Drugs 13:1925–1965
Deutch CE (1994) Characterization of a novel salt-tolerant Bacillus sp. from the nasal cavity of desert iguanas. FEMS Microbiol Lett 121:55–60
Deutch CE (2002) Characterization of a salt-tolerant extracellular α-amylase from Bacillus dipsosauri. Lett Appl Microbiol 35:78–84
Diop A, Khelaifia S, Armstrong N, Labs N, Fourrnier PE, Raoult D, Million M (2016) Microbial culturonics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb Ecol Health Dis 27:32049. https://doi.org/10.3402/mehd.v27.32049
Diop A, Seck E, Dubourg G, Armstrong N, Blanc-Tailleur C, Raoult D, Fournier PE (2018) Genome sequence and description of Gracilibacillus timonensis sp. nov. strain Marseille- P2481T, a moderate halophilic bacterium isolated from the human gut microflora. MicrobiologyOpen 8:e638. https://doi.org/10.1002/mbo3.638
Duchêne D, Bochot A (2016) Thirty years of cyclodextrins. Int J Pharm 514:58–72
Editorial Board (2015) Gracilibacillus. Bergey’s Manual of Systematics of Archaea and Bacteria Online. https://doi.org/10.1002/9781118960608.gbm00534
Ekblom R, Wolf JBW (2014) A field guide to whole genome sequencing, assembly and annotation. Evol Appl 7:1026–1042
Foo AY, Bais R (1998) Amylase measurement with 2-chloro-4-nitrophenyl-maltotrioside as substrate. Clin Chim Acta 272:137–147
Gao M, Li ZZ, Zhou YG, Liu HC, Ma YC, Wang L, Chen SF, Ji XC (2012) Gracilibacillus kekensis sp. nov., a moderate halophile isolated from Keke Salt Lake. Int J Syst Evol Microbiol 62:1032–1036
Gella FJ, Gubern G, Vidal R, Canalias F (1997) Determination of total and pancreatic α-amylase in human serum with 2-chloro-4-nitrophenyl-α-d-maltotrioside as substrate. Clinica Chemica Acta 259:147–160
Gerday C, Glansdorff N (2007) Physiology and biochemistry of extremophiles. ASM Press, Washington
Goel A, Nene SN (1993) Modifications in the phenolphthalein method for spectrophotometric estimation of beta cyclodextrin. Starch (Stäke) 47:399–400
Graziano G, Merlino A (2014) Molecular basis of protein halotolerance. Biochim Biophys Acta 1844:850–858
Guan TW, Tian L, Li EY, Tang SK, Zhang XP (2017) Gracilibacillus aidingensis sp. nov., a novel moderately halophilic bacterium isolated from Aiding salt lake. Arch Microbiol 199:1277–1281
Guan HL, Zhang YJ, Lu XJ, Jia M, Zhang ZY, Gao XH, Ma YC, Tian F, Tang SK (2018) Gracilibacillus eburneus sp. nov., a moderately halophilic bacterium isolated form Xinjiang province, China. Archiv Microbiol 200:423–429
Hames BD (1998) Gel electrophoresis of proteins: a practical approach (3/e). Oxford University Press, Oxford
Hirota K, Hanaoka Y, Nodasaka Y, Yumoto I (2014) Gracilibacillus alcaliphilus sp. nov., a facultative alkaliphile isolated from indigo fermentation liquor for dyeing. Int J Syst Evol Microbiol 64:3174–3180
Horikoshi K, Grant WD (1998) Extremophiles: microbial life in extreme environments. Wiley-Liss, New York
Hough DW, Danson MJ (1999) Extremozymes. Curr Opin Chem Biol 3:39–46
Huang HQ, Wang Y, Yuan WD, Xiao C, Ye JJ, Liu M, Zhu J, Sun QG, Bao SX (2013) Gracilibacillus marinus sp. nov., isolated from the northern South China Sea. Antonie Van Leeuwenhoek 104:695–701
Huo YY, Xu XW, Cui HL, Wu M (2010) Gracilibacillus ureilyticus sp. nov., a halotolerant bacterium from a saline-alkaline soil. Int J Syst Evol Microbiol 60:1383–1386
Jansook P, Ogawa N, Loftsson T (2018) Cyclodextrins: structure, physicochemical properties and pharmacological applications. Int J Pharm 535:272–284
Jeon CO, Lim JM, Jang HH, Park DJ, Xu LH, Jiang CL, Kim CJ (2008) Gracilibacillus lacisalsi sp. nov., a halophilic Gram-positive bacterium from a salt lake in China. Int J Sys Evol Microbiol 58:2282–2286
Kato T, Horikoshi K (1984) Colorimetric determination of γ-cyclodextrin. Anal Chem 56:1738–1740
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858
Kim P, Lee JC, Park DJ, Shin KS, Kim JY, Kim CJ (2012) Gracilibacillus bigeumensis sp. nov., a moderately halophilic bacterium from solar saltern soil. Int J Syst Evol Microbiol 62:1857–1863
Krüger A, Schäfers C, Schröder C, Antranikian G (2018) Towards a sustainable biobased industry—highlighting the impact of extremophiles. New Biotechnol 40:144–153
Laderman KA, Davis BR, Krutzsch HC, Lewis MS, Griko YV, Privalov PL, Anfinsen CB (1993) The purification and characterization of an extremely thermostable α-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Biol Chem 268:24394–24401
Lanyi JK (1974) Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev 38:272–290
Lawson PA, Deutch CE, Collins MD (1996) Phylogenetic characterization of a novel salt- tolerant Bacillus species: description of Bacillus dipsosauri sp. nov. J Appl Bacteriol 81:109–112
Leemhuis H, Kelly RM, Dijkhuizen L (2010) Engineering of cyclodextrin glucanotransferases and the impact for biotechnological applications. Appl Microbiol Biotechnol 85:823–835
Lejeune A, Sakaguchi K, Imanaka T (1989) A spectrophotometric assay for the cyclization activity of cyclomaltohexaose (α-cyclodextrin) glucanotransferase. Anal Biochem 181:6–11
Lorentz K, Gütschow B, Renner F (1999) Evaluation of direct α-amylase assay using 2-chloro-4- nitrophenyl-α-d-maltotrioside. Clin Chem Lab Med 37:1053–1062
Madern D, Ebel C, Zaccai G (2000) Halophilic adaptation of enzymes. Extremophiles 4:91–98
Madigan MT, Marrs BL (1997) Extremophiles. Sci Am 276:82–87
McCleary BV (1988) Soluble dye-labeled polysaccharides for the assay of endohydrolases. Methods Enzymol 160:74–86
Merino N, Aronson HS, Bojanova DP, Feyhl-Buska J, Wong ML, Zhang S, Giovannelli D (2019) Living at the extremes: extremophiles and the limits of life in a planetary context. Front Microbiol 10:780. https://doi.org/10.3389/fmicb.2019.00780
Niehaus F, Bertoldo C, Kähler Antrankian G (1999) Extremophiles as a source of novel enzymes for industrial application. Appl Microbiol Biotechnol 51:711–729
Oh YJ, Lee HW, Kim SK, Kwon MS, Lee J, Jang JY, Park HW, Nam YD, Seo MJ, Choi HJ (2016) Gracilibacillus kimchii sp. nov., a halophilic bacterium isolated from kimchi. J. Microbiol 54:588–593
Orellana R, Macaya C, Bravo G, Dorochesi F, Cumsille A, Valencia R, Rojas C, Seeger M (2018) Living at the frontiers of life: extremophiles in Chile and their potential for bioremediation. Front Microbiol 9:2309. https://doi.org/10.3369/fmicb.2018.02309
Qi Q, Zimmermann W (2005) Cyclodextrin glucanotransferase: from gene to applications. Appl Microbiol Biotechnol 66:474–484
Rothschild LJ, Mancinelli RL (2001) Life in extreme environments Nature 409:1092–1101
Schiraldi C, De Rosa M (2002) The production of biocatalysts and biomolecules from extremophiles. Trends Biotechnol 20:515–521
Senghor B, Khelaifia S, Bassêne H, Seck EH, Fournier PE, Sokhna C, Raoult D, Lagier JC (2017) ‘Gracilibacillus phocaeensis’ sp. nov., ‘Sediminibacillus massiliensis’ sp. nov., and ‘Virgibacillus ndiopensis’ sp. nov., three halophilic species isolated from salty human stools by culturomics. New Microb New Infect 20:51–54
Sheehan H, McCleary BV (1988) A new procedure for the measurement of fungal and bacterial α-amylase. Biotechnol Tech 2:289–292
Simpson JT, Pop M (2015) The theory and practice of genome sequence assembly. Ann Rev Genomcs Hum Genet 16:151–172
Smith P, Krohn RI, Hermanson GT, Malina K, Garner FH, Provensano MD, Fugimoto EK, Goeke NM, Olson GJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85
Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B (2006) Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel 19:555–562
Suganuma T, Maeda Y, Kitahara K, Nagahama T (1997) Study of the action of human salivary alpha-amylase on 2-chloro-4-nitrophenyl-α-maltotrioside in the presence of potassium thiocyanate. Carbohydr Res 303:219–227
Tang SK, Wang Y, Lou K, Mao PH, Jin X, Jiang CL, Xu LH, Li WJ (2009) Gracilibacillus saliphilus sp. nov., a moderately halophilic bacterium isolated from a salt lake. Int J Syst Evol Microbiol 59:1620–1624
Wainø M, Tindall BJ, Schumann P, Ingvorsen K (1999) Gracilibacillus gen. nov., with description of Gracilibacillus halotolerans gen. nov., sp. nov.; transfer of Bacillus dipsosauri to Gracilibacillus dipsosauri comb. nov., and Bacillus salexigens to the genus Salibacillus gen. nov., as Salibacillus salexigens comb. nov. Int J Syst Bacteriol 49:821–831
Winn-Deen ES, David H, Sigler G, Chavez R (1988) Development of a direct assay for α- amylase. Clin Chem 34:2005–2008
Yang N, Ren B, Dai H, Liu Z, Zhou Y, Song F, Zhang L (2013) Gracilibacillus xinjiangesnsis sp. nov., a new member of the genus Gracilibacillus isolated from the Xinjiang region, China. Antonie Van Leeuwenhoek 104:809–816
Acknowledgements
We thank Maria Dolores Sotelo for her efforts to clone the α-amylase gene from G. dipsosauri and Drs. Pamela Marshall and Jeffrey Newman for facilitating the sequencing of the G. dipsosauri genome at Indiana University.
Funding
There was no external funding source for this work.
Author information
Authors and Affiliations
Contributions
Charles E. Deutch did all of the microbiological and biochemical experiments and wrote the manuscript. Shanshan Yang did the assembly and analysis of the Gracilibacillus dipsosauri genome.
Corresponding author
Ethics declarations
Conflicts of interest
We declare that we have no conflicts of interest in publishing this work.
Human or animal participants
No humans or animals were used in this project.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Multiple sequence alignment of the α-amylases from G. dipsosauri (PWU69658.1), G. halophilus (N4WMC6), G. orientalis (A0A1H9MMU5), G. kekensis (A0A1M7K410), and G. ureilyticus (A0A1I4H7R9). Fully conserved residues are indicated by (*); similar residues are indicated by (:) or (.) (PNG 146 kb)
Rights and permissions
About this article
Cite this article
Deutch, C.E., Yang, S. Genomic sequencing of Gracilibacillus dipsosauri reveals key properties of a salt-tolerant α-amylase. Antonie van Leeuwenhoek 113, 1049–1059 (2020). https://doi.org/10.1007/s10482-020-01417-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01417-2