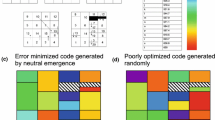
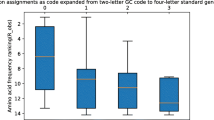
Abstract
Stereochemical nucleotide-amino acid interactions, in the form of noncovalent nucleotide-amino acid interactions, potentially produced the genetic code’s codon-amino acid assignments. Empirical estimates of single nucleotide-amino acid affinities on surfaces and in solution are used to test whether trinucleotide-amino acid affinities determined genetic code assignments pending the principle “first arrived, first served”: presumed early amino acids have greater codon-amino acid affinities than ulterior ones. Here, these single nucleotide affinities are used to approximate all 64 × 20 trinucleotide-amino acid affinities. Analyses show that (1) on surfaces, genetic code codon-amino acid assignments tend to match high affinities for the amino acids that integrated earliest the genetic code (according to Wong’s metabolic coevolution hypothesis between nucleotides and amino acids) and (2) in solution, the same principle holds for the anticodon-amino acid assignments. Affinity analyses match best genetic code assignments when assuming that trinucleotides competed for amino acids, rather than amino acids for trinucleotides. Codon-amino acid affinities stick better to genetic code assignments than anticodon-amino acid affinities. Presumably, two independent coding systems, on surfaces and in solution, converged, and formed the current translation system. Proto-translation on surfaces by direct codon-amino acid interactions without tRNA-like adaptors coadapted with a system emerging in solution by proto-tRNA anticodon-amino acid interactions. These systems assigned identical or similar cognates to codons on surfaces and to anticodons in solution. Results indicate that a prebiotic metabolism predated genetic code self-organization.


Similar content being viewed by others
Data availability
All disclosed in tables and supplement.
References
Abrahão J, Silva L, Silva LS, Khalil JYB, Rodrigues R, Arantes T, Assis F, Boratto P, Andrade M, Kroon EG, Ribeiro B, Bergier I, Seligmann H, Ghigo E, Colson P, Levasseur A, Kroemer G, Raoult D, LaScola B (2018) Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat Commun 9:749
Agmon I (2009) The dimeric proto-ribosome: structural details and possible implications on the origin of life. Int J Mol Sci 10:2921–2934
Agmon IC (2016) Could a proto-ribosome emerge spontaneously in the prebiotic world? Molecules 21:e1701
Ahmed A, Frey G, Michel CJ (2007) Frameshift signals in genes associated with the circular code. In Silico Biol 7:155–168
Ahmed A, Frey G, Michel CJ (2010) Essential molecular functions associated with the circular code evolution. J Theor Biol 264:613–622
Amunts A, Brown A, Bai XC, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V (2014) Structure of the yeast mitochondrial large ribosomal subunit. Science 343:1485–1489
Ardell DH (1998) On error minimization in a sequential origin of the standard genetic code. J Mol Evol 47:1–13
Arquès DG, Michel CJ (1996) A complementary circular code in the protein coding genes. J Theor Biol 182:45–58
Barthélémy RM, Seligmann H (2016) Cryptic tRNAs in chaetognath mitochondrial genomes. Comput Biol Chem 95:119–132
Bartonek L, Zagrovic B (2017) mRNA/protein sequence complementarity and its determinants: the impact of affinity scales. PLoS Comput Biol 13:e1005648
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300
Błazej P, Wnetrzak M, Mackiewicz D, Mackiewicz P (2019) The influence of different types of translational inaccuracies on the genetic code structure. BMC Bioinformatics 20:114
Bloch DP, McArthur B, Widdowson R, Spector D, Guimarães RC, Smith J (1983) tRNA-rRNA sequence homologies: evidence for a common evolutionary origin? J Mol Evol 19:420–428
Bloch DP, McArthur B, Widdowson R, Spector D, Guimarães RC, Smith J (1984) tRNA-rRNA sequence homologies: a model for the origin of a common ancestral molecule, and prospects for its reconstruction. Orig Life 14:571–578
Bloch DP, McArthur B, Guimarães RC, Smith J, Staves MP (1989) tRNA-rRNA sequence matches from inter- and intraspecies comparisons suggest common origins for the two RNAs. Braz J Med Biol Res 22:931–944
Brown A, Rathore S, Kimanius D, Aibara S, Bai XC, Rorbach J, Amunts A, Ramakrishnan V (2017) Structures of the human mitochondrial ribosome in native states of assembly. Nat Struct Mol Biol 24:866–869
Caetano-Anollés G (2002) Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Res 30(11):2575–2587
Caetano-Anollés G, Caetano-Anollés D (2015a) Computing the origin and evolution of the ribosome from its structure – uncovering processes of macromolecular accretion benefiting synthetic biology. Comput Struct Biotechnol J 13:427–447
Caetano-Anollés D, Caetano-Anollés G (2015b) Ribosomal accretion, apriorism and the phylogenetic method: a response to Petrov and Williams. Front Genet 6:194
Caetano-Anollés D, Caetano-Anollés G (2016a) Piecemeal buildup of the genetic code, ribosomes, genomes from primordial tRNA building blocks. Life (Basel) 6:e43
Caetano-Anollés D, Caetano-Anollés G (2016b) Commentary: history of the ribosome and the origin of translation. Front Mol Biosci 3:87
Caetano-Anollés G, Sun F-J (2014) The natural history of transfer RNA and its interactions with the ribosome. Front Genet 5:127
Caetano-Anollés G, Kim HS, Mittenthal JE (2007) The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci U S A 104:9358–9363
Caetano-Anollés G, Yafremava LS, Gee H, Caetano-Anollés D, Kim HS, Mittenthal JE (2009) The origin and evolution of modern metabolism. Int J Biochem Cell Biol 41:285–297
Caporaso JG, Yarus M, Knight R (2005) Error minimization and coding triplet/binding site associations are independent features of the canonical genetic code. J Mol Evol 61:597–607
Caudron B, Jestin JL (2012) Sequence criteria for the anti-parallel character of protein betastrands. J Theor Biol 315:146–149
Curgy JJ (1985) The mitoribosomes. Biol Cell 54:1–38
Demongeot J, Moreira A (2007) A possible circular RNA at the origin of life. J Theor Biol 249(2):314–324
Demongeot J, Norris V (2019) Emergence of a “cyclosome” in a primitive network capable of building “infinite” proteins. Life (Basel) 6:e51
Demongeot J, Seligmann H (2019a) Spontaneous evolution of circular codes in theoretical minimal RNA rings. Gene 705:95–102
Demongeot J, Seligmann H (2019b) Theoretical minimal RNA rings recapitulate the order of the genetic code's codon-amino acid assignments. J Theor Biol 471:108–116
Demongeot J, Seligmann H (2019c) Bias for 3′-dominant codon directional asymmetry in theoretical minimal RNA rings. J Comput Biol 26:1003–1012
Demongeot J, Seligmann H (2019d) More pieces of ancient than recent theoretical minimal proto-tRNA-like RNA rings in genes coding for tRNA synthetases. J Mol Evol 87:152–174
Demongeot J, Seligmann H (2019e) Theoretical minimal RNA rings designed according to coding constraints mimick deamination gradients. Naturwissenschaften 106:44
Demongeot J, Seligmann H (2019f) The Uroboros theory of life’s origin: 22-nucleotide theoretical minimal RNA rings reflect evolution of genetic code and tRNA-rRNA translation machineries. Acta Biotheor 67:273–297
Demongeot J, Seligmann H (2019g) Evolution of tRNA into rRNA secondary structures. Gene Rep 17:100483
Demongeot J, Seligmann H (2020a) Pentamers with non-redundant frames: bias for natural circular code codons. J Mol Evol 88:194–201
Demongeot J, Seligmann H (2020b) The primordial tRNA acceptor stem code from theoretical minimal RNA ring clusters. BMC Genet 21:7
Demongeot J, Seligmann H (2020c) RNA rings strengthen hairpin accretion hypotheses for tRNA evolution: a reply to commentaries by Z.F. Burton and M. Di Giulio, J Mol Evol in press
Demongeot J, Seligmann H (2020d) Accretion history of large ribosomal subunits deduced from theoretical minimal RNA rings is congruent with histories derived from phylogenetic and structural methods. Gene 738:144438
Demongeot J, Seligmann H (2020e) Comparisons between small ribosomal RNA and theoretical minimal RNA ring secondary structures confirm phylogenetic and structural accretion histories. Sci Rep (in press)
Demongeot J, Seligmann H (2020f) Theoretical minimal RNA rings mimic molecular evolution before tRNA-mediated translation: codon-amino acid affinities increase from early to late RNA rings. C R Biol (in press)
Demongeot J, Seligmann H (2020g) Deamination gradients within codons after 1<−>2 position swap predict amino acid hydrophobicity and parallel β-sheet conformational preference. Biosystems 191-192:104116
Du X, Xia Y-L, Ai S-M, Liang J, Ji X-L, Liu S-Q (2016) Insights into protein-ligand interactions: mechanisms, models, and methods. Int J Mol Sci 17:144
Farias ST, Rêgo TG, José MV (2014) Origin and evolution of the peptidyl transferase center from proto-tRNAs. FEBS Open Bio 4:175–178
Farias ST, Rêgo TG, José MV (2019) Origin of the 16S ribosomal molecular from ancestral tRNAs. Sci 1:8
Faure E, Barthélémy R-M (2018) True mitochondrial tRNA punctuation and initiation using overlapping stop and start codons at specific and conserved positions. In Mitochondrial DNA; Seligmann H and Warthi G (eds), IntechOpen, London, 2018, 3–29
Faure E, Barthélémy R-M (2019) Specific mitochondrial ss-tRNAs in phylum Chaetognatha. J Entomol Zool Stud 7:304–315
Faure E, Delaye L, Tribolo S, Levasseur A, Seligmann H, Barthélémy RM (2011) Probable presence of an ubiquitous cryptic mitochondrial gene on the antisense strand of the cytochrome oxidase I gene. Biol Direct 6:56
Ferus M, Pietrucci F, Saitta AM, Knožek A, Kubelik P, Ivanek O, Shestivska V, Civiš S (2017) Formation of nucleobases in a Miller-Urey reducing atmosphere. Proc Natl Acad Sci U S A 114:4306–4311
Fimmel E, Michel CJ, Starman M, Strüngmann L (2018) Self-complementary circular codes in coding theory. Theory Biosci 137:51–85
Fontecilla-Camps JC (2014) The stereochemical basis of the genetic code and the (mostly) autotrophic origin of life. Life (Basel) 4:1013–1025
Fontecilla-Camps JC (2019) Geochemical continuity and catalyst/cofactor replacement in the emergence and evolution of life. Angew Chem Int Ed Eng 58:42–48
Freeland SJ, Hurst LD (1998) The genetic code is one in a million. J Mol Evol 47:238–248
Geyer R, Madany MA (2018) On the efficiency of the genetic code after frameshift mutations. PeerJ 6:e4825
Goldford JE, Segré D (2018) Modern views of ancient metabolic networks. Curr Op Syst Biol 8:117–124
Griffith RW (2009) A specific scenario for the origin of life and the genetic code based on peptide/oligonucleotide interdependence. Orig Life Evol Biosph 39:517–531
Guilloux A, Jestin JL (2012) The genetic code and its optimization for kinetic energy conservation in polypeptide chains. Biosystems 109:141–144
Guimarães RC (2011) Metabolic basis for the self-referential genetic code. Orig Life Evol Biosph 41:357–371
Guimarães RC (2015) The self-referential genetic code is biologic and includes the error minimization property. Orig Life Evol Biosph 45:69–75
Guimarães RC (2017) Self-referential encoding on modules of anticodon pairs—roots of the biological flow system. Life 7:16
Guimarães RC, Moreira CH, de Farias ST (2008) A self-referential model for the formation of the genetic code. Theory Biosci 127:249–270
Han DX, Wang HY, Ji ZL, Hu AF, Zhao YF (2010) Amino acid homochirality may be linked to the origin of phosphate-based life. J Mol Evol 70:572–582
Harish A, Caetano-Anollés G (2012) Ribosomal history reveals origins of modern protein synthesis. PLoS One 7:e32776
Hartman H (1975a) Speculations on the evolution of the genetic code. Orig Life 6:423–427
Hartman H (1975b) Speculations on the evolution of the genetic code. J Mol Evol 4:359–370
Hartman H (1978) Speculations on the evolution of the genetic code. II. Orig Life 9:133–136
Hartman H (1995) Speculations on the evolution of the genetic code. J Mol Evol 40:541–544
Hartman H (1998) Photosynthesis and the origin of life. Orig Life Evol Biosph 28:515–521
Hartman H, Smith TF (2017) The evolution of the ribosome and the genetic code. Life (Basel) 20:227–249
Hobish MK, Wickramasinghe NSMD, Ponnamperuma C (1995) Direct interactions between amino acids and nucleotides as a possible physicochemical basis for the origin of the genetic code. Adv Space Res 15:368–382
Hörst SM, Yelle RV, Buch A, Carrasco N, Cernogora G, Dutuit O, Quirico E, Sciamma O'BE, Smith MA, Somogyi A, Szopa C, Thissen R, Vuitton V (2012) Formation of amino acids and nucleotide bases in a titan atmosphere simulation experiment. Astrobiology 12:809–817
Ilardo M, Meringer M, Freeland S, Rasulev B, Cleaves HJ II (2015) Extraordinarily adaptive properties of the genetically encoded amino acids. Sci Rep 5:9414
Janas T, Janas T, Yarus M (2006) Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res 34:2128–2136
Johnson DBF, Wang L (2010) Imprints of the genetic code in the ribosome. Proc Natl Acad Sci U S A 107:8298–8303
Kumar B, Saini S (2016) Analysis of the optimality of the standard genetic code. Mol BioSyst 12:2642–2651
Lancet D, Zidovetzki R, Markovitch O (2018) Systems protobiology: origin of life in lipid catalytic networks. J R Soc Interface 15:20180159
Michel CJ (2012) Circular code motifs in transfer and 16S ribosomal RNAs: a possible translation code in genes. Comput Biol Chem 37:24–37
Michel CJ (2013) Circular code motifs in transfer RNAs. Comput Biol Chem 45:17–29
Michel CJ (2017) The maximal C3 self-complementary trinucleotide circular code X in genes of bacteria, archaea, eukaryotes, plasmids and viruses. Life (Basel) 7:e20
Michel CJ, Seligmann H (2014) Bijective transformation circular codes and nucleotide exchanging RNA transcription. Biosystems 118:39–50
Miller SL (1953) Production of amino acids under possible primitive earth conditions. Science 117:528–529
Miller SL, Urey HC (1959) Organic compound synthesis on the primitive earth. Science 130:254–251
Miyazawa S, Jernigan RL (1985) Estimation of effective interresidue contact energies from protein crystal structures: quasi-chemical approximation. Macromolecules 18:534–552
Miyazawa S, Jernigan RL (1997) Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term. J Mol Biol 256:623–644
Möller W, Janssen GM (1990) Transfer RNAs for primordial amino acids contain remnants of a primitive code at position 3 to 5. Biochimie 72:361–368
Möller W, Janssen GM (1992) Statistical evidence for remnants of the primordial code in the acceptor stem of prokaryotic transfer RNA. J Mol Evol 34:471–477
Moraveç J, El Din SB, Seligmann H, Sivan N, Werner YL (1999) Systematics and distribution of the Acanthodactylus pardalis group (Reptilia: Sauria: Lacertidae) in Egypt and Israel. Zoology in the Middle East 17:21–50
Nasir A, Caetano-Anollés G (2015) A phylogenomic data-driven exploration of viral origins and evolution. Sci Adv 1:e15000527
Nasir A, Kim KM, Caetano-Anollés G (2017) Phylogenetic tracings of proteome size support the gradual accretion of protein structural domains and the early origin of viruses from primordial cells. Front Microbiol 8:1178
Pelc SR (1965) Correlation between coding-triplets and amino acids. Nature 207:597–599
Pelc SR, Welton MGE (1966) Stereochemical relationship between coding triplets and aminoacids. Nature 209:868–870
Perneger TV (1998) What’s wrong with Bonferroni adjustments. BMJ 316:1236–1238
Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD (2015) History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A 112:15396–15401
Philip GK, Freeland SJ (2011) Did evolution select a nonrandom “alphabet” of amino acids? Astrobiology 11:235–240
Polyansky AA, Zagrovic B (2013) Evidence of direct complementary interactions between messenger RNAs and their cognate proteins. Nucleic Acids Res 41:83434–88443
Raiser M (2018) An appeal to magic? The discovery of a non-enzymatic metabolism and its role in the origins of life. Biochem J 475:2577–2592
Rocha EPC (2004) Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res 14:2279–2286
Rogers SO (2019) Evolution of the genetic code based on conservative changes of codons, amino acids, and aminoacyl tRNA synthetases. J Theor Biol 466:1–10
Root-Bernstein R (2007) Simultaneous origin of homochirality, the genetic code and its directionality. BioEssays 29:689–698
Root-Bernstein R (2010) Experimental test of L- and D-amino acid binding to L- and D-codons suggests that homochirality and codon directionality emerged with the genetic code. Symmetry 2:1180–1200
Seligmann H (1998) Evidence that minor directional asymmetry is functional in lizard hindlimbs. J Zool (Lond) 248:205–208
Seligmann H (2000) Evolution and ecology of developmental processes and of the resulting morphology: directional asymmetry in hindlimbs of Agamidae and Lacertidae (Reptilia: Lacertilia). Biol J Linn Soc 69:461–481
Seligmann H (2002) Behavioural and morphological asymmetries in hindlimbs of Hoplodactylus duvaucelii (Lacertilia : Gekkonomorpha : Gekkota : Diplodactylinae). Laterality 7:277–283
Seligmann H (2007) Cost minimization of ribosomal frameshifts. J Theor Biol 249:162–167
Seligmann H (2010a) Do anticodons of misacylated tRNAs preferentially mismatch codons coding for the misloaded amino acid? BMC Mol Biol 11:41
Seligmann H (2010b) The ambush hypothesis at the whole-organism level: off frame, 'hidden' stops in vertebrate mitochondrial genes increase developmental stability. Comput Biol Chem 35:81–95
Seligmann H (2010c) Undetected antisense tRNAs in mitochondrial genomes? Biol Direct 5:39
Seligmann H (2010d) Avoidance of antisense, antiterminator tRNA anticodons in vertebrate mitochondria. Biosystems 101:42–50
Seligmann H (2011a) Error compensation of tRNA misacylation by codon-anticodon mismatch prevents translational amino acid misinsertion. Comput Biol Chem 35:81–95
Seligmann H (2011b) Two genetic codes, one genome: frameshifted primate mitochondrial genes code for additional proteins in presence of antisense antitermination tRNAs. Biosystems 105:271–285
Seligmann H (2011c) An overlapping genetic code for frameshifted overlapping genes in Drosophila mitochondria: antisense antitermination tRNAs UAR insert serine. J Theor Biol 298:51–76
Seligmann H (2012a) Coding constraints modulate chemically spontaneous mutational replication gradients in mitochondrial genomes. Curr Genomics 13:37–54
Seligmann H (2012b) Overlapping genetic codes for overlapping frameshifted genes in Testudines, and Lepidochelys olivacea as special case. Comput Biol Chem 41:18–34
Seligmann H (2015a) Phylogeny of genetic codes and punctuation codes within genetic codes. Biosystems 129:36–43
Seligmann H (2015b) Codon expansion and systematic transcriptional deletions produce tetra-, pentacoded mitochondrial peptides. J Theor Biol 387:154–165
Seligmann H (2016a) Translation of mitochondrial swinger RNAs according to tri-, tetra- and pentacodons. Biosystems 140:38–48
Seligmann H (2016b) Unbiased mitoproteome analyses confirm non-canonical RNA, expanded codon translations. Comp Struct Biotechnol J 14:391–403
Seligmann H (2016c) Natural chymotrypsin-like-cleaved human mitochondrial peptides confirm tetra-, pentacodon, non-canonical RNA translations. Biosystems 147:78–93
Seligmann H (2017a) Natural mitochondrial proteolysis confirms transcription systematically exchanging/deleting nucleotides, peptides coded by expanded codons. J Theor Biol 414:76–90
Seligmann H (2017b) Reviewing evidence for systematic transcriptional deletions, nucleotide exchanges, and expanded codons, and peptide clusters in human mitochondria. Biosystems 160:10–24
Seligmann H (2018a) Protein sequences recapitulate genetic code evolution. Comp Struct Biotech J 16:177–199
Seligmann H (2018b) Giant viruses as protein-coated mitochondria? Virus Res 253:77–86
Seligmann H (2018c) Alignment-based and alignment-free methods converge with experimental data on amino acids coded by stop codons at split between nuclear and mitochondrial genetic codes. Biosystems 167:33–46
Seligmann H (2018d) Directed mutations recode mitochondrial genes: from regular to stoplessgenetic codes. Chapter in: mitochondrial DNA, Seligmann H and Warthi G eds, InTechOpen
Seligmann H (2019a) Localized context-dependent effects of the “ambush” hypothesis: more off-frame stop codons downstream of shifty codons. DNA Cell Biol 38:786–795
Seligmann H (2019b) Giant viruses: spore-like missing links between rickettsia and mitochondria? Ann N Y Acad Sci 1447:69–79
Seligmann H (2019c) Syntenies between cohosted mitochondrial, chloroplast and Phycodnavirus genomes: functional mimicry and/or common ancestry? DNA Cell Biol 38:1257–1268
Seligmann H, Amzallag GN (2002) Chemical interactions between amino acid and RNA: multiplicity of the levels of specificity explains origin of the genetic code. Naturwissenschaften 89:542–551
Seligmann H, Demongeot J (2020) Codon directional asymmetry suggests swapped prebiotic 1st and 2nd codon positions. Int J Mol Sci 21:E347
Seligmann H, Labra A (2013) Tetracoding increases with body temperature in Lepidosauria. Biosystems 114:155–163
Seligmann H, Pollock DD (2004) The ambush hypothesis: hidden stop codons prevent offframe gene reading. DNA Cell Biol 23:707–714
Seligmann H, Raoult D (2016) Unifying view of stem-loop hairpin RNA as origin of current and ancient parasitic and non-parasitic RNAs, including in giant viruses. Curr Opin Microbiol 31:1–8
Seligmann H, Raoult D (2018) Stem-loop RNA hairpins in giant viruses: invading rRNA-like repeats and a template free RNA. Front Microbiol 9:101
Seligmann H, Warthi G (2017) Genetic code optimization for cotranslational protein folding: codon directional asymmetry correlates with antiparallel betasheets, tRNA synthetase classes. Comp Struct Biotech J 48:412–424
Seligmann H, Beiles A, Werner YL (2003a) More injuries in left-footed lizards. J Zool (Lond) 260:129–144
Seligmann H, Beiles A, Werner YL (2003b) Avoiding injury or adapting to survive injury? Two coexisting strategies in lizards. Biol J Linn Soc 78:307–324
Seligmann H, Moravec J, Werner YL (2008) Morphological, functional and evolutionary aspects of tail autotomy and regeneration in the “living fossil” Sphenodon (Reptilia: Rhynchocephalia). Biol J Linn Soc 93:721–743
Szathmáry E (1993) Coding coenzyme handles: a hypothesis for the origin of the genetic code. Proc Natl Acad Sci U S A 90:9916–9920
Trifonov EN (2000) Consensus temporal order of amino acids and evolution of the triplet code. Gene 261:139–151
Trifonov EN (2004) The triplet code from first principles. J Biomol Struct Dyn 22:1–11
Turk-Macleod RM, Puthenvedu D, Majerfeld I, Yarus M (2012) The plausibility of RNA-templated peptides: simultaneous RNA affinity for adjacent peptide side chains. J Mol Evol 61:226–235
Wang X, Dong Q, Chen G, Zhang J, Liu Y, Zhao J, Peng H, Wang Y, Cai Y, Wang X, Yang C (2016) The universal genetic code, protein coding genes and genomes of all species were optimized for frameshift tolerance. bioRxiv. https://doi.org/10.1101/067736
Weber AL, Lacey JC Jr (1978) Genetic code correlations: amino acids and their anticodon nucleotides. J Mol Evol 11:199–210
Woese CR (1965) Order in the genetic code. Proc Nat Acad Sci U S A 54:71–75
Wong JTF (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci 72:1909–1912
Wong JTF (2005) The coevolution hypothesis at age thirty. Bioessays 27:416–426
Yarus M (2017) The genetic code and RNA-amino acid affinities. Life (Basel, Switzerland) 7:e13
Yarus M, Christian EL (1989) Genetic code origins. Nature 342:349–350
Yarus M, Widmann JJ, Knight R (2009) RNA-amino acid binding: a stereochemical era for the genetic code. J Mol Evol 69:406–429
Zagrovic B, Bartonek L, Polyansky AA (2018) RNA-protein interactions in an unstructured context. FEBS Lett 592:2901–2916
Acknowledgments
Thanks are due to Francisco Prosdocimi and two anonymous reviewers for improving previous versions.
Author information
Authors and Affiliations
Contributions
HS developed ideas, gathered, and analyzed data and wrote as a sole author of this study.
Corresponding author
Ethics declarations
Competing interests
The author declares that he has no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Additional information
Communicated by: William Benjamin Walker
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 34 kb)
Rights and permissions
About this article
Cite this article
Seligmann, H. First arrived, first served: competition between codons for codon-amino acid stereochemical interactions determined early genetic code assignments. Sci Nat 107, 20 (2020). https://doi.org/10.1007/s00114-020-01676-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00114-020-01676-z