Abstract
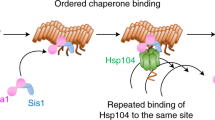
Amyloid appearance is a rare event that is promoted in the presence of other aggregated proteins. These aggregates were thought to act by templating the formation of an assembly-competent nucleation seed, but we find an unanticipated role for them in enhancing the persistence of amyloid after it arises. Specifically, Saccharomyces cerevisiae Rnq1 amyloid reduces chaperone-mediated disassembly of Sup35 amyloid, promoting its persistence in yeast. Mathematical modeling and corresponding in vivo experiments link amyloid persistence to the conformationally defined size of the Sup35 nucleation seed and suggest that amyloid is actively cleared by disassembly below this threshold to suppress appearance of the [PSI+] prion in vivo. Remarkably, this framework resolves multiple known inconsistencies in the appearance and curing of yeast prions. Thus, our observations establish the size of the nucleation seed as a previously unappreciated characteristic of prion variants that is key to understanding transitions between prion states.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
Code for the stochastic model of aggregate persistence is available at https://doi.org/10.6071/M33T08
References
Tuite, M. F. & Serio, T. R. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat. Rev. Mol. Cell Biol. 11, 823–833 (2010).
Knowles, T. P. J., Vendruscolo, M. & Dobson, C. M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014).
Serio, T. R. [PIN +]ing down the mechanism of prion appearance. FEMS Yeast Res. 18, 924 (2018).
Masel, J., Jansen, V. A. & Nowak, M. A. Quantifying the kinetic parameters of prion replication. Biophys. Chem. 77, 139–152 (1999).
Masel, J. & Jansen, V. A. The measured level of prion infectivity varies in a predictable way according to the aggregation state of the infectious agent. Biochim. Biophys. Acta 1535, 164–173 (2001).
Knowles, T. P. et al. An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009).
Gaspar, R. et al. Secondary nucleation of monomers on fibril surface dominates α-synuclein aggregation and provides autocatalytic amyloid amplification. Q. Rev. Biophys. 50, e6 (2017).
Jarrett, J. T. & Lansbury, P. T. Seeding ‘one-dimensional crystallization’ of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 73, 1055–1058 (1993).
Come, J. H., Fraser, P. E. & Lansbury, P. T. A kinetic model for amyloid formation in the prion diseases: importance of seeding. Proc. Natl Acad. Sci. USA 90, 5959–5963 (1993).
Nowak, M. A., Krakauer, D. C., Klug, A. & May, R. M. Prion infection dynamics. Integr. Biol. 1, 3–15 (1998).
Powers, E. T., Morimoto, R. I., Dillin, A., Kelly, J. W. & Balch, W. E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991 (2009).
Koga, H., Kaushik, S. & Cuervo, A. M. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res. Rev. 10, 205–215 (2011).
Derkatch, I. L., Bradley, M. E., Hong, J. Y. & Liebman, S. W. Prions affect the appearance of other prions: the story of [PIN +]. Cell 106, 171–182 (2001).
Osherovich, L. Z. & Weissman, J. S. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI +] prion. Cell 106, 183–194 (2001).
Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R. & Morimoto, R. I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 (2006).
Tyedmers, J., Madariaga, M. L. & Lindquist, S. Prion switching in response to environmental stress. PLoS Biol. 6, e294 (2008).
Cox, B. S. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity 20, 505–521 (1965).
Doel, S. M., McCready, S. J., Nierras, C. R. & Cox, B. S. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137, 659–670 (1994).
Ter-Avanesyan, M. D., Dagkesamanskaya, A. R., Kushnirov, V. V. & Smirnov, V. N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [PSI +] in the yeast Saccharomyces cerevisiae. Genetics 137, 671–676 (1994).
Chernoff, Y. O., Lindquist, S. L., Ono, B., Inge-Vechtomov, S. G. & Liebman, S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI +]. Science 268, 880–884 (1995).
Patino, M. M., Liu, J. J., Glover, J. R. & Lindquist, S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273, 622–626 (1996).
Paushkin, S. V., Kushnirov, V. V. & Smirnov, V. N. Propagation of the yeast prion-like [PSI +] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134 (1996).
Glover, J. R. et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI +], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 (1997).
King, C. Y. et al. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl Acad. Sci. USA 94, 6618–6622 (1997).
Paushkin, S. V., Kushnirov, V. V., Smirnov, V. N. & Ter-Avanesyan, M. D. In vitro propagation of the prion-like state of yeast Sup35 protein. Science 277, 381–383 (1997).
Derkatch, I. L., Bradley, M. E., Zhou, P., Chernoff, Y. O. & Liebman, S. W. Genetic and environmental factors affecting the de novo appearance of the [PSI +] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 (1997).
Sarell, C. J., Stockley, P. G. & Radford, S. E. Assessing the causes and consequences of co-polymerization in amyloid formation. Prion 7, 359–368 (2013).
Krebs, M. R. H., Morozova-Roche, L. A., Daniel, K. & Dobson, C. M. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci. 13, 1933–1938 (2004).
Resende, C. et al. The Candida albicans Sup35p protein (CaSup35p): function, prion-like behaviour and an associated polyglutamine length polymorphism. Microbiology 148, 1049–1060 (2002).
Santoso, A., Chien, P., Osherovich, L. Z. & Weissman, J. S. Molecular basis of a yeast prion species barrier. Cell 100, 277–288 (2000).
Tanaka, M., Collins, S. R., Toyama, B. H. & Weissman, J. S. The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 (2006).
Derdowski, A., Sindi, S. S., Klaips, C. L., DiSalvo, S. & Serio, T. R. A size threshold limits prion transmission and establishes phenotypic diversity. Science 330, 680–683 (2010).
Dickinson, A. G., Fraser, H., Meikle, V. M. & Outram, G. W. Competition between different scrapie agents in mice. Nat. New Biol. 237, 244–245 (1972).
Dickinson, A. G. Host–pathogen interactions in scrapie. Genetics 79 (Suppl.), 387–395 (1975).
Manuelidis, L. Vaccination with an attenuated Creutzfeldt–Jakob disease strain prevents expression of a virulent agent. Proc. Natl Acad. Sci. USA 95, 2520–2525 (1998).
Bartz, J. C. et al. Prion interference is due to a reduction in strain-specific PrPSc levels. J. Virol. 81, 689–697 (2007).
Tanaka, M. et al. Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004).
King, C.-Y. Transformation of yeast by infectious prion particles. Methods 39, 68–71 (2006).
Song, Y. et al. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 4, 289–297 (2005).
Satpute-Krishnan, P., Langseth, S. X. & Serio, T. R. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 5, e24 (2007).
Higurashi, T., Hines, J. K., Sahi, C., Aron, R. & Craig, E. A. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl Acad. Sci. USA 105, 16596–16601 (2008).
Tipton, K. A., Verges, K. J. & Weissman, J. S. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell 32, 584–591 (2008).
Pei, F., DiSalvo, S., Sindi, S. S. & Serio, T. R. A dominant-negative mutant inhibits multiple prion variants through a common mechanism. PLoS Genet. 13, e1007085 (2017).
DiSalvo, S., Derdowski, A., Pezza, J. A. & Serio, T. R. Dominant prion mutants induce curing through pathways that promote chaperone-mediated disaggregation. Nat. Struct. Mol. Biol. 18, 486–492 (2011).
Klaips, C. L., Hochstrasser, M. L., Langlois, C. R. & Serio, T. R. Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. Elife 3, 1751 (2014).
Sondheimer, N. & Lindquist, S. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5, 163–172 (2000).
Derkatch, I. L., Chernoff, Y. O., Kushnirov, V. V., Inge-Vechtomov, S. G. & Liebman, S. W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 (1996).
Bagriantsev, S. & Liebman, S. W. Specificity of prion assembly in vivo. [PSI +] and [PIN +] form separate structures in yeast. J. Biol. Chem. 279, 51042–51048 (2004).
Lancaster, A. K., Bardill, J. P., True, H. L. & Masel, J. The spontaneous appearance rate of the yeast prion [PSI +] and its implications for the evolution of the evolvability properties of the [PSI +] system. Genetics 184, 393–400 (2010).
Zhou, P., Derkatch, I. L. & Liebman, S. W. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI +] and [PIN +]. Mol. Microbiol. 39, 37–46 (2001).
Borchsenius, A. S., Muller, S., Newnam, G. P., Inge-Vechtomov, S. G. & Chernoff, Y. O. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr. Genet. 49, 21–29 (2006).
Kryndushkin, D. S., Engel, A., Edskes, H. & Wickner, R. B. Molecular chaperone Hsp104 can promote yeast prion generation. Genetics 188, 339–348 (2011).
Pezza, J. A., Villali, J. & Serio, T. R. Amyloid-associated activity contributes to the severity and toxicity of a prion phenotype. Nat. Commun. 5, 4384 (2014).
Sharma, J. & Liebman, S. W. Exploring the basis of [PIN +] variant differences in [PSI +] induction. J. Mol. Biol. 425, 3046–3059 (2013).
Gillespie, D. T. The chemical Langevin equation. J. Chem. Phys. 113, 297–306 (2000).
Ferrone, F. A., Hofrichter, J. & Eaton, W. A. Kinetics of sickle hemoglobin polymerization. II. A double nucleation mechanism. J. Mol. Biol. 183, 611–631 (1985).
Wegrzyn, R. D., Bapat, K., Newnam, G. P., Zink, A. D. & Chernoff, Y. O. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell Biol. 21, 4656–4669 (2001).
Kryndushkin, D. S., Ter-Avanesyan, M. D. & Kushnirov, V. V. Yeast [PSI +] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278, 49636–49643 (2003).
Liu, J. J. & Lindquist, S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature 400, 573–576 (1999).
Langlois, C. R., Pei, F., Sindi, S. S. & Serio, T. R. Distinct prion domain sequences ensure efficient amyloid propagation by promoting chaperone binding or processing in vivo. PLoS Genet. 12, e1006417 (2016).
Hofrichter, J., Ross, P. D. & Eaton, W. A. Kinetics and mechanism of deoxyhemoglobin S gelation: a new approach to understanding sickle cell disease. Proc. Natl Acad. Sci. USA 71, 4864–4868 (1974).
Yang, Z., Hong, J. Y., Derkatch, I. L. & Liebman, S. W. Heterologous Gln/Asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet. 9, e1003236 (2013).
Arslan, F., Hong, J. Y., Kanneganti, V., Park, S.-K. & Liebman, S. W. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genet. 11, e1004814 (2015).
Bradley, M. E., Edskes, H. K., Hong, J. Y., Wickner, R. B. & Liebman, S. W. Interactions among prions and prion ‘strains’ in yeast. Proc. Natl Acad. Sci. USA 99 (Suppl. 4), 16392–16399 (2002).
Mathur, V., Hong, J. Y. & Liebman, S. W. Ssa1 overexpression and [PIN +] variants cure [PSI +] by dilution of aggregates. J. Mol. Biol. 390, 155–167 (2009).
Westergard, L. & True, H. L. Extracellular environment modulates the formation and propagation of particular amyloid structures. Mol. Microbiol. 92, 698–715 (2014).
Khan, T. et al. Quantifying nucleation in vivo reveals the physical basis of prion-like phase behavior. Mol. Cell 71, 155–168 (2018).
Park, K.-W., Hahn, J.-S., Fan, Q., Thiele, D. J. & Li, L. De novo appearance and ‘strain’ formation of yeast prion [PSI +] are regulated by the heat-shock transcription factor. Genetics 173, 35–47 (2006).
Stein, K. C. & True, H. L. Extensive diversity of prion strains is defined by differential chaperone interactions and distinct amyloidogenic regions. PLoS Genet. 10, e1004337 (2014).
Dulle, J. E. & True, H. L. Low activity of select Hsp104 mutants is sufficient to propagate unstable prion variants. Prion 7, 394–403 (2013).
Dulle, J. E., Stein, K. C. & True, H. L. Regulation of the Hsp104 middle domain activity is critical for yeast prion propagation. PLoS ONE 9, e87521 (2014).
Bradley, M. E. & Liebman, S. W. Destabilizing interactions among [PSI +] and [PIN +] yeast prion variants. Genetics 165, 1675–1685 (2003).
Stein, K. C., Bengoechea, R., Harms, M. B., Weihl, C. C. & True, H. L. Myopathy-causing mutations in an HSP40 chaperone disrupt processing of specific client conformers. J. Biol. Chem. 289, 21120–21130 (2014).
Zampieri, M., Legname, G. & Altafini, C. Investigating the conformational stability of prion strains through a kinetic replication model. PLoS Comput. Biol. 5, e1000420 (2009).
Sun, Y. et al. Conformational stability of PrP amyloid fibrils controls their smallest possible fragment size. J. Mol. Biol. 376, 1155–1167 (2008).
Gao, X. et al. Human Hsp70 disaggregase reverses Parkinson’s-linked α-synuclein amyloid fibrils. Mol. Cell 59, 781–793 (2015).
Kirstein, J. et al. In vivo properties of the disaggregase function of J-proteins and Hsc70 in Caenorhabditis elegans stress and aging. Aging Cell 16, 1414–1424 (2017).
Serio, T. R., Cashikar, A. G., Moslehi, J. J., Kowal, A. S. & Lindquist, S. L. Yeast prion [PSI +] and its determinant, Sup35p. Methods Enzymol. 309, 649–673 (1999).
Tanaka, M. A protein transformation protocol for introducing yeast prion particles into yeast. Methods Enzymol. 470, 681–693 (2010).
Pezza, J. A. et al. The NatA acetyltransferase couples Sup35 prion complexes to the [PSI +] phenotype. Mol. Biol. Cell 20, 1068–1080 (2009).
Satpute-Krishnan, P. & Serio, T. R. Prion protein remodelling confers an immediate phenotypic switch. Nature 437, 262–265 (2005).
Kushnirov, V. V., Alexandrov, I. M., Mitkevich, O. V., Shkundina, I. S. & Ter-Avanesyan, M. D. Purification and analysis of prion and amyloid aggregates. Methods 39, 50–55 (2006).
Acknowledgements
We thank S. Lindquist (MIT) and E. Craig (University of Wisconsin–Madison) for reagents and J. Laney and members of the Serio and Sindi groups for helpful discussions and comments on the manuscript. This research was sponsored in part by the Joint DMS/NIGMS Initiative to Support Research at the Interface of the Biological and Mathematical Sciences (R01GM126548 to S.S.S.), NSF-INSPIRE (1344279 to S.S.S. and J.K.D.) and NIH/NIGMS (F32GM096582 to J.V., R35GM118042 to T.R.S.) and with support from NIH/NIGMS to T.M.B. (T32GM008659).
Author information
Authors and Affiliations
Contributions
J.V. and J.D. conceptualized and designed the work, acquired, analyzed and interpreted data, and drafted and revised the manuscript. T.M.B. and F.P. acquired, analyzed and interpreted data and revised the manuscript. S.S.S. and T.R.S. conceptualized and designed the work, analyzed and interpreted data, and drafted and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Protein abundance for strains with heterozygous disruptions of HSP104 and/or RNQ1.
The relative levels of Hsp104 (a), Sup35 (b), and Rnq1 (c) in lysates from diploid yeast strains analyzed in Fig. 1e–g were determined by SDS-PAGE and immunoblotting with specific antisera. Strains carried wildtype (+/+), heterozygous (+/Δ), or homozygous (Δ/Δ) disruptions of HSP104 or RNQ1 and the indicated variant of [PSI+]. Protein levels are expressed relative to the protein levels in wildtype version of the same prion strain (that is [PSI+]Strong or [PSI+]Weak) in arbitrary units (a.u.). Data are mean and s.d. from 3 independent experiments performed with independent cultures. Data for the graphs in panels a-b are available as source data.
Extended Data Fig. 2 Hsp104 abundance, availability and association with Sup35 in strains with varying copy number or propagating [PIN+].
The relative levels of Hsp104 (a) and Sup35 (b) in lysates from diploid yeast strains used to determine the frequency of [PSI+] appearance in response to transient overexpression of the Sup35 prion domain (Fig. 2c) were determined by SDS-PAGE and immunoblotting with specific antisera. Strains carried wildtype (+/+), heterozygous disruptions of HSP104 (+/Δ), or an extra copy of HSP104 (+/++). Protein levels are expressed relative to wildtype in arbitrary units (a.u.). Data are mean and s.d. from 3 independent experiments performed with independent cultures. c, The frequency of spontaneous loss of the indicated [PSI+] variant was determined in yeast strains carrying wildtype (+/+, black), heterozygous disruptions of HSP104 (+/Δ, gray), or an extra copy of HSP104 (+/++). Data shown are means; error bars represent 95% confidence intervals from ten independent cultures. d, Representative immunoblots of SDS-PAGE gels following immunocapture of NM-HA from lysates isolated from the indicated [PSI+] variant using either specific (anti-HA) or non-specific (anti-MYC) beads. Both [PIN+] (+) and [pin−] (-) strains were analyzed for capture of NM-HA and co-capture of Sup35 and Hsp104, using specific anti-sera. A non-specific band cross-reacting with the HA antiserum is indicated (*). Data for the graphs in panels a-c and uncropped images for panel d are available as source data.
Extended Data Fig. 3 Aggregate amplification probabilities and size distributions.
A stochastic persistence model of Sup35 aggregate dynamics in vivo determines the probability of amplification of a single Sup35 aggregate as a function of fragmentation rate with Sup35 expression levels (a) at 25% (red), 50% (orange), 100% (teal), 200% (blue) or 400% (purple) or Sup35 aggregate numbers (b) of one (red), two (orange), four (blue) or eight (purple) for [PSI+]Strong with a seed size of five. c, Aggregate size distributions for [PSI+]Strong with a seed size of five (solid black), [PSI+]Weak with a seed size of five (dashed red), or [PSI+]Weak with a seed size of fifteen (solid red) are shown.
Extended Data Fig. 4 Aggregate size distribution shifts in response to experimental perturbation.
The aggregate size distributions revealed by our deterministic model (shaded) shift in response to inhibition of Sup35 synthesis (a, b) or Hsp104-mediated fragmentation (c, d) after 90 min (solid, unshaded),180 min (dashed, unshaded), 270 min (dotted, unshaded) or at its steady-state (gray, unshaded) for strains propagating [PSI+]Strong with a seed size of 5 (a, c) or [PSI+]Weak with a seed size of fifteen (b, d). e, The distribution of aggregate sizes before (Xi, solid black) and after (Ci(t), dashed black) t minutes of inhibition of Sup35 synthesis is shown. The shift in the size distribution, S(t), is quantified as the area between the curves when Ci(t) > Xi (shaded area). f, The size distribution of Sup35 aggregates from a strain propagating [PSI+]Weak with a seed size of five (shaded) shifts upon inhibition of Sup35 synthesis for 90 min (solid, unshaded), 180 min (dashed, unshaded), 270 min (dotted, unshaded) or at its steady-state (gray, unshaded). g, and h, The shifts in aggregate size distribution over a range of fragmentation and conversion rates that match the steady-state soluble level of Sup35 were monitored at 90 min (solid), 180 min (dashed), 270 min (dotted) and steady-state (gray) for a strain propagating [PSI+]Weak with a seed size of five (g) or fifteen (h). The x-axis shows the fragmentation rate scaled to the strain-specific value in Supplementary Table 2 (vertical black line). While [PSI+]Weak with a seed size of five (g) shifts slowly to its steady-state distribution, [PSI+]Weak with a seed size of fifteen (h) rapidly reaches is steady-state distribution, matching our empirical observations (Fig. 4a).
Extended Data Fig. 5 Fiber transfection fitting.
Fiber transfection experiments from Fig. 1 were fit mathematically to determine the number of fibers required to induce a stable [PSI+] state. Shown are the observations (black) and fits for fibers of the prion domain of Sup35 assembled at 4 °C (Sc4) or at 37 °C (Sc37) in [pin-] (a, c) or [PIN+] (b, d) yeast strains to a model of one (red), two (green), or three (blue) fibers. Data shown are means and s.d. from at least two independent experiments performed with independent cultures.
Extended Data Fig. 6 Schematics of mathematical models.
a, Schematic of nucleated polymerization dynamics used in the mathematical models. Non-prion state Sup35 (circle) is synthesized at rate α and can join either end an amyloid aggregate of Sup35 (polymer of squares) at a conversion rate β. Amyloid aggregates are fragmented at rate γ at the interface between any two monomers. If the resulting two aggregates are both greater in size than the minimum seed (ns), the amyloid state persists (blue); if either aggregate is smaller in size than ns, it will disassemble into non-prion state Sup35 (red). b, The stochastic persistence model calculates the probability of each of three outcomes, relative to cell division, upon the introduction of a single aggregate of minimal size under nucleated polymerization dynamics: aggregate disassembly, retention of a single aggregate, and aggregate persistence (that is the creation of two aggregates greater in size than ns).
Supplementary information
Supplementary Information
Supplementary Note 1 (mathematical supplement) and Supplementary Tables 1–5.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Uncropped western blot.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Uncropped western blot.
Rights and permissions
About this article
Cite this article
Villali, J., Dark, J., Brechtel, T.M. et al. Nucleation seed size determines amyloid clearance and establishes a barrier to prion appearance in yeast. Nat Struct Mol Biol 27, 540–549 (2020). https://doi.org/10.1038/s41594-020-0416-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0416-6