Abstract
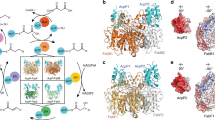
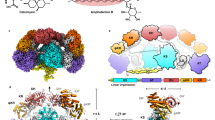
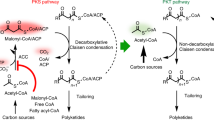
In type II polyketide synthases (PKSs), the ketosynthase–chain length factor (KS–CLF) complex catalyzes polyketide chain elongation with the acyl carrier protein (ACP). Highly reducing type II PKSs, represented by IgaPKS, produce polyene structures instead of the well-known aromatic skeletons. Here, we report the crystal structures of the Iga11–Iga12 (KS–CLF) heterodimer and the covalently cross-linked Iga10=Iga11–Iga12 (ACP=KS–CLF) tripartite complex. The latter structure revealed the molecular basis of the interaction between Iga10 and Iga11–Iga12, which differs from that between the ACP and KS of Escherichia coli fatty acid synthase. Furthermore, the reaction pocket structure and site-directed mutagenesis revealed that the negative charge of Asp 113 of Iga11 prevents further condensation using a β-ketoacyl product as a substrate, which distinguishes IgaPKS from typical type II PKSs. This work will facilitate the future rational design of PKSs.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Klaus, M. & Grininger, M. Engineering strategies for rational polyketide synthase design. Nat. Prod. Rep. 35, 1070–1081 (2018).
Staunton, J. & Weissman, K. J. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18, 380–416 (2001).
Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716 (2009).
Hertweck, C., Luzhetskyy, A., Rebets, Y. & Bechthold, A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24, 162–190 (2007).
Das, A. & Khosla, C. Biosynthesis of aromatic polyketides in bacteria. Acc. Chem. Res. 42, 631–639 (2009).
Tsai, S.-C. The structural enzymology of iterative aromatic polyketide synthases: a critical comparison with fatty acid synthases. Annu. Rev. Biochem. 87, 503–531 (2018).
Dreier, J. & Khosla, C. Mechanistic analysis of a type II polyketide synthase. Role of conserved residues in the β-ketoacyl synthase-chain length factor heterodimer. Biochemistry 39, 2088–2095 (2000).
Du, D. et al. Production of a novel amide-containing polyene by activating a cryptic biosynthetic gene cluster in Streptomyces sp. MSC090213JE08. ChemBioChem 17, 1464–1471 (2016).
Du, D., Katsuyama, Y., Shin-Ya, K. & Ohnishi, Y. Reconstitution of a type II polyketide synthase that catalyzes polyene formation. Angew. Chem. Int. Ed. Engl. 57, 1954–1957 (2018).
Grammbitter, G. L. C. et al. An uncommon type II PKS catalyzes biosynthesis of aryl polyene pigments. J. Am. Chem. Soc. 141, 16615–16623 (2019).
Keatinge-Clay, A. T., Maltby, D. A., Medzihradszky, K. F., Khosla, C. & Stroud, R. M. An antibiotic factory caught in action. Nat. Struct. Mol. Biol. 11, 888–893 (2004).
Shen, H. C. et al. Synthesis and biological evaluation of platensimycin analogs. Bioorg. Med. Chem. Lett. 19, 1623–1627 (2009).
Trajtenberg, F. et al. Structural insights into bacterial resistance to cerulenin. FEBS J. 281, 2324–2338 (2014).
Wang, J. et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441, 358–361 (2006).
Nanson, J. D., Himiari, Z., Swarbrick, C. M. D. & Forwood, J. K. Structural characterisation of the beta-ketoacyl-acyl carrier protein synthases, FabF and FabH, of Yersinia pestis. Sci. Rep. 5, 14797 (2015).
Zhang, Y.-M., Hurlbert, J., White, S. W. & Rock, C. O. Roles of the active site water, histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae. J. Biol. Chem. 281, 17390–17399 (2006).
Moche, M., Schneider, G., Edwards, P., Dehesh, K. & Lindqvist, Y. Structure of the complex between the antibiotic cerulenin and its target, β-ketoacyl-acyl carrier protein synthase. J. Biol. Chem. 274, 6031–6034 (1999).
Finzel, K., Lee, D. J. & Burkart, M. D. Using modern tools to probe the structure-function relationship of fatty acid synthases. Chembiochem 16, 528–547 (2015).
Crosby, J. & Crump, M. P. The structural role of the carrier protein–active controller or passive carrier. Nat. Prod. Rep. 29, 1111–1137 (2012).
Shakya, G. et al. Modeling linear and cyclic PKS intermediates through atom replacement. J. Am. Chem. Soc. 136, 16792–16799 (2014).
Chen, A., Re, R. N. & Burkart, M. D. Type II fatty acid and polyketide synthases: deciphering protein-protein and protein-substrate interactions. Nat. Prod. Rep. 35, 1029–1045 (2018).
Miyanaga, A., Iwasawa, S., Shinohara, Y., Kudo, F. & Eguchi, T. Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis. Proc. Natl Acad. Sci. USA 113, 1802–1807 (2016).
Miyanaga, A. et al. Structural basis of protein-protein interactions between a trans-acting acyltransferase and acyl carrier protein in polyketide disorazole biosynthesis. J. Am. Chem. Soc. 140, 7970–7978 (2018).
Nguyen, C. et al. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature 505, 427–431 (2014).
Milligan, J. C. et al. Molecular basis for interactions between an acyl carrier protein and a ketosynthase. Nat. Chem. Biol. 15, 669–671 (2019).
Dodge, G. J. et al. Structural and dynamical rationale for fatty acid unsaturation in Escherichia coli. Proc. Natl Acad. Sci. USA 116, 6775–6783 (2019).
Herbst, D. A. et al. The structural organization of substrate loading in iterative polyketide synthases. Nat. Chem. Biol. 14, 474–479 (2018).
Worthington, A. S., Rivera, H., Torpey, J. W., Alexander, M. D. & Burkart, M. D. Mechanism-based protein cross-linking probes to investigate carrier protein-mediated biosynthesis. ACS Chem. Biol. 1, 687–691 (2006).
Worthington, A. S. & Burkart, M. D. One-pot chemo-enzymatic synthesis of reporter-modified proteins. Org. Biomol. Chem. 4, 44–46 (2006).
Clarke, K. M., Mercer, A. C., La Clair, J. J. & Burkart, M. D. In vivo reporter labeling of proteins via metabolic delivery of coenzyme A analogues. J. Am. Chem. Soc. 127, 11234–11235 (2005).
Haushalter, R. W., Worthington, A. S., Hur, G. H. & Burkart, M. D. An orthogonal purification strategy for isolating crosslinked domains of modular synthases. Bioorg. Med. Chem. Lett. 18, 3039–3042 (2008).
Worthington, A. S. et al. Probing the compatibility of type II ketosynthase-carrier protein partners. Chembiochem 9, 2096–2103 (2008).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Acknowledgements
We thank A. Harada and M. Senda, and the staff of the Photon Factory and Swiss Light Source (proposal number 2017G165) for the X-ray data collection. The research is supported by Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (JP18H02144 and JP19H04645 to Y.K.), a Research Fellow Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS; JP17J09439, to D.D.), JSPS A3 Foresight Program grant (to Y.O.), NIH R01 grant GM095970 to M.D.B. and NIH K12 GM068524 grant, to T.D.D. (San Diego IRACDA Postdoctoral Fellow).
Author information
Authors and Affiliations
Contributions
Y.K., M.D.B. and D.D. designed the research. T.D.D. synthesized the chemical probes. The crystallization of the proteins was done by D.D. The X-ray diffraction experiment was carried out by Y.K. and D.D. The experimental phasing was done by Y.K. Refinement and validation of the structure was done by D.D., Y.K. and S.F. Site-directed mutagenesis and analysis of mutants were done by D.D. and M.H. The cross-linked complex was prepared by D.D. and A.C. D.D. and Y.K. wrote the draft manuscript. A.C., T.D.D., M.D.B., S.F. and Y.O. commented on the draft. Y.K. and Y.O. finalized the manuscript and all authors approved it. Y.K., M.D.B. and Y.O. directed the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplemental Information
Supplementary Figs. 1–14 and Tables 1–4.
Rights and permissions
About this article
Cite this article
Du, D., Katsuyama, Y., Horiuchi, M. et al. Structural basis for selectivity in a highly reducing type II polyketide synthase. Nat Chem Biol 16, 776–782 (2020). https://doi.org/10.1038/s41589-020-0530-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0530-0
This article is cited by
-
Discovery of type II polyketide synthase-like enzymes for the biosynthesis of cispentacin
Nature Communications (2023)
-
Enzymology of assembly line synthesis by modular polyketide synthases
Nature Chemical Biology (2023)
-
C–N bond formation by a polyketide synthase
Nature Communications (2023)
-
Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis
Scientific Reports (2021)