Abstract
Purpose
To investigate the between-laboratory reproducibility of embryo selection/deselection effectiveness using qualitative and quantitative time-lapse parameters.
Methods
A systematic search was performed on MEDLINE, EMBASE, and the Cochrane Library (up to February 2020) without restriction on date, language, document type, and publication status. Measuring outcomes included implantation, blastulation, good-quality blastocyst formation, and euploid blastocyst.
Results
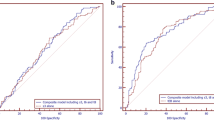
We detected 6 retrospective cohort studies externally validating the first clinical time-lapse model (Meseguer) emphasizing quantitative parameters, of which 3 (including one involving 2 independent centers) were included for the pooled analysis. Receiver operating characteristics analysis showed reduced predictive power of the model when either including or not including sister clinic validation. Fifteen cohort studies evaluating qualitative parameters were included for meta-analysis, and the mean Newcastle-Ottawa Scale was 5.3. Overall, meta-analysis showed significantly adverse association between the presence of ≥ 1 cleavage abnormalities and embryo implantation rates (11 studies, n = 7266; RR = 0.39[0.28, 0.55]95% CI; I2 = 57%). Further analysis showed adverse impacts of direct cleavage (7 studies, n = 7065; RR = 0.28 [0.15, 0.54] 95% CI; I2 = 46%), reverse cleavage (2 studies, n = 3622; RR = 0.16 [0.03, 0.75] 95% CI; I2 = 0%), chaotic cleavage (2 studies, n = 3643; RR = 0.11 [0.02, 0.69] 95% CI; I2 = 24%), and multinucleation (5 studies, n = 2576; RR = 0.59 [0.50, 0.69] 95% CI; I2 = 0%), but not the < 6 intercellular contact points at the 4-cell stage (1 study, n = 185; RR = 0.17 [0.02, 1.15] 95% CI).
Conclusions
Qualitative time-lapse parameters are reliably associated with embryo developmental potential among laboratories, whereas the reproducibility of time-lapse embryo selection model that emphasizes quantitative parameters may be compromised when externally applied.




Similar content being viewed by others
References
Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–71.
Basile N, Vime P, Florensa M, Aparicio Ruiz B, Garcia Velasco JA, Remohi J, et al. The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Hum Reprod. 2015;30:276–83.
Liu Y, Chapple V, Feenan K, Roberts P, Matson P. Time-lapse deselection model for human day 3 in vitro fertilization embryos: the combination of qualitative and quantitative measures of embryo growth. Fertil Steril. 2016;105:656–62 e651.
Milewski R, Kuc P, Kuczynska A, Stankiewicz B, Lukaszuk K, Kuczynski W. A predictive model for blastocyst formation based on morphokinetic parameters in time-lapse monitoring of embryo development. J Assist Reprod Genet. 2015;32:571–9.
Petersen BM, Boel M, Montag M, Gardner DK. Development of a generally applicable morphokinetic algorithm capable of predicting the implantation potential of embryos transferred on day 3. Hum Reprod. 2016;31:2231–44.
Freour T, Le Fleuter N, Lammers J, Splingart C, Reignier A, Barriere P. External validation of a time-lapse prediction model. Fertil Steril. 2015;103:917–22.
Liu Y, Feenan K, Chapple V, Matson P. Assessing efficacy of day 3 embryo time-lapse algorithms retrospectively: impacts of dataset type and confounding factors. Hum Fertil (Camb). 2019;22:182–90.
Yalcinkaya E, Ergin EG, Caliskan E, Oztel Z, Ozay A, Ozornek H. Reproducibility of a time-lapse embryo selection model based on morphokinetic data in a sequential culture media setting. J Turk Ger Gynecol Assoc. 2014;15:156–60.
Kirkegaard K, Hindkjaer JJ, Ingerslev HJ. Effect of oxygen concentration on human embryo development evaluated by time-lapse monitoring. Fertil Steril. 2013;99:738–44 e734.
Wissing ML, Bjerge MR, Olesen AIG, Hoest T, Mikkelsen AL. Impact of PCOS on early embryo cleavage kinetics. Reprod BioMed Online. 2014;28:508–14.
Rubio I, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escriba MJ, et al. Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil Steril. 2012;98:1458–63.
Liu Y, Chapple V, Roberts P, Matson P. Prevalence, consequence, and significance of reverse cleavage by human embryos viewed with the use of the Embryoscope time-lapse video system. Fertil Steril. 2014;102:1295–1300.e1292.
Athayde Wirka K, Chen AA, Conaghan J, Ivani K, Gvakharia M, Behr B, et al. Atypical embryo phenotypes identified by time-lapse microscopy: high prevalence and association with embryo development. Fertil Steril. 2014, 1637-1648(101):e1631–5.
Liu Y, Chapple V, Feenan K, Roberts P, Matson P. Clinical significance of intercellular contact at the four-cell stage of human embryos, and the use of abnormal cleavage patterns to identify embryos with low implantation potential: a time-lapse study. Fertil Steril. 2015;103:1485–1491.e1481.
Ergin EG, Caliskan E, Yalcinkaya E, Oztel Z, Cokelez K, Ozay A, et al. Frequency of embryo multinucleation detected by time-lapse system and its impact on pregnancy outcome. Fertil Steril. 2014;102:1029–1033.e1021.
Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73:1155–8.
Higgins JPT GSe. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 [updated March 2011]. The Cochrane Collaboration. 2011.
Wells GASB, O’Connell D, Peterson J, Welch V, et al. The Newcastle -Ottawa scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. 2011. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 19 Oct 2019
Team RC.R. A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2017.
Rubio I, Galan A, Larreategui Z, Ayerdi F, Bellver J, Herrero J, et al. Clinical validation of embryo culture and selection by morphokinetic analysis: a randomized, controlled trial of the EmbryoScope. Fertil Steril. 2014;102:1287–1294.e1285.
Desch L, Bruno C, Luu M, Barberet J, Choux C, Lamotte M, et al. Embryo multinucleation at the two-cell stage is an independent predictor of intracytoplasmic sperm injection outcomes. Fertil Steril. 2017;107:97–103.e104.
Balakier H, Sojecki A, Motamedi G, Librach C. Impact of multinucleated blastomeres on embryo developmental competence, morphokinetics, and aneuploidy. Fertil Steril. 2016;106:608–614.e602.
Barrie A, Homburg R, McDowell G, Brown J, Kingsland C, Troup S. Preliminary investigation of the prevalence and implantation potential of abnormal embryonic phenotypes assessed using time-lapse imaging. Reprod BioMed Online. 2017;34:455–62.
Desai N, Goldberg JM, Austin C, Falcone T. Are cleavage anomalies, multinucleation, or specific cell cycle kinetics observed with time-lapse imaging predictive of embryo developmental capacity or ploidy? Fertil Steril. 2018;109:665–74.
Fan YL, Han SB, Wu LH, Wang YP, Huang GN. Abnormally cleaving embryos are able to produce live births: a time-lapse study. J Assist Reprod Genet. 2016;33:379–85.
Hashimoto S, Nakano T, Yamagata K, Inoue M, Morimoto Y, Nakaoka Y. Multinucleation per se is not always sufficient as a marker of abnormality to decide against transferring human embryos. Fertil Steril. 2016;106:133–139.e136.
Hojnik N, Vlaisavljevic V, Kovacic B. Morphokinetic characteristics and developmental potential of in vitro cultured embryos from natural cycles in patients with poor ovarian response. Biomed Res Int. 2016; 2016:4286528.
Hur YS, Ryu EK, Hyun CS, Yang SH, Yoon SH, Lim KS, et al. Retrospective study of single vitrified-warmed blastocyst transfer cycles according to the presence of morphokinetic variables. Clin Exp Reprod Med. 2018;45:52–5.
Yang ST, Shi JX, Gong F, Zhang SP, Lu CF, Tan K, et al. Cleavage pattern predicts developmental potential of day 3 human embryos produced by IVF. Reprod BioMed Online. 2015;30:625–34.
Almagor M, Or Y, Fieldust S, Shoham Z. Irregular cleavage of early preimplantation human embryos: characteristics of patients and pregnancy outcomes. J Assist Reprod Genet. 2015;32:1811–5.
Kirkegaard K, Kesmodel US, Hindkjaer JJ, Ingerslev HJ. Time-lapse parameters as predictors of blastocyst development and pregnancy outcome in embryos from good prognosis patients: a prospective cohort study. Hum Reprod. 2013;28:2643–51.
Kong X, Yang S, Gong F, Lu C, Zhang S, Lu G, et al. The relationship between cell number, division behavior and developmental potential of cleavage stage human embryos: a time-lapse study. PLoS ONE [Electronic Resource]. 2016;11:e0153697.
Yang SH, Wu CH, Chen YC, Yang CK, Wu TH, Chen PC, et al. Effect of morphokinetics and morphological dynamics of cleavage stage on embryo developmental potential: a time-lapse study. Taiwan J Obstet Gynecol. 2018;57:76–82.
Lagalla C, Tarozzi N, Sciajno R, Wells D, Di Santo M, Nadalini M, et al. Embryos with morphokinetic abnormalities may develop into euploid blastocysts. Reprod BioMed Online. 2017;34:137–46.
Armstrong S, Bhide P, Jordan V, Pacey A, Farquhar C. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane Database Syst Rev. 2018;5:CD011320.
Pribenszky C, Nilselid AM, Montag M. Time-lapse culture with morphokinetic embryo selection improves pregnancy and live birth chances and reduces early pregnancy loss: a meta-analysis. Reprod BioMed Online. 2017;35:511–20.
Liu Y, Copeland C, Stevens A, Feenan K, Chapple V, Myssonski K, et al. Assessment of human embryos by time-lapse videography: a comparison of quantitative and qualitative measures between two independent laboratories. Reprod Biol. 2015;15:210–6.
Ciray HN, Aksoy T, Goktas C, Ozturk B, Bahceci M. Time-lapse evaluation of human embryo development in single versus sequential culture media--a sibling oocyte study. J Assist Reprod Genet. 2012;29:891–900.
Ciray HN, Campbell A, Agerholm IE, Aguilar J, Chamayou S, Esbert M, et al. Proposed guidelines on the nomenclature and annotation of dynamic human embryo monitoring by a time-lapse user group. Hum Reprod. 2014;29:2650–60.
Liu Y, Sakkas D, Afnan M, Matson P. Time-lapse videography for embryo selection/de-selection: a bright future or fading star? Hum Fertil (Camb). 2019; 9:1–7.
Fishel S, Campbell A, Montgomery S, Smith R, Nice L, Duffy S, et al. Time-lapse imaging algorithms rank human preimplantation embryos according to the probability of live birth. Reprod BioMed Online. 2018;37:304–13.
Tran D, Cooke S, Illingworth PJ, Gardner DK. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum Reprod. 2019;34:1011–8.
Vayena E, Blasimme A, Cohen IG. Machine learning in medicine: addressing ethical challenges. PLoS Med. 2018;15:e1002689.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplemental Table 1
Characteristics of included studies for quantitative assessment (external validation of the Meseguer model) (DOCX 14 kb).
Supplemental Table 2
Characteristics of included studies for qualitative assessment (DOCX 17 kb).
Supplemental File 1
(DOCX 33 kb).
Rights and permissions
About this article
Cite this article
Liu, Y., Qi, F., Matson, P. et al. Between-laboratory reproducibility of time-lapse embryo selection using qualitative and quantitative parameters: a systematic review and meta-analysis. J Assist Reprod Genet 37, 1295–1302 (2020). https://doi.org/10.1007/s10815-020-01789-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-020-01789-4