Abstract
Increasing temperatures on a global scale and locally deteriorating water quality affect coral distribution and health. Mechanisms that convey environmental robustness are poorly understood and have been attributed to the coral host, algal symbionts, and prokaryotic associates. Flexibility of the host’s (bacterial) microbiome has been suggested to contribute to environmental robustness, but the underlying mechanisms are unclear. We therefore utilised the vastly contrasting water quality gradient present along Hong Kong’s highly urbanised coastline to explore whether flexibility in the microbiome of Oulastrea crispata relates to spatial variations in temperature, salinity, dissolved oxygen, pH, nitrate, nitrite, ammonia, total nitrogen, phosphorus, turbidity, and chlorophyll a. We identified differences in the coral microbiomes between sites, but the measured environmental variables only explained ~ 23% of the variation suggesting other factors are contributing substantially. The observed structural complexity of the microbiome (based on alpha diversity indices) appears to be relatively conserved across the environmental gradient even at sites where no other hard coral can survive. Therefore, we conclude that, at least in O. crispata, flexibility in the microbiome does not appear to underpin the robustness of this broadly distributed coral.
Similar content being viewed by others
Introduction
Human impacts lead to both global- and local-scale environmental changes and cause the ongoing decline of coral reefs documented through the Anthropocene (Hughes et al. 2017). Stressors including increasing sea surface temperatures and physical habitat destruction may completely extirpate most corals before the end of the century (Hoegh-Guldberg et al. 2018). Ultimately, worldwide action to decrease the CO2 emissions that drive global change is essential to preserve the ecologic and economic services of reefs. Under less extreme conditions, variation in coral’s stress response (colony to colony and species to species) and mortality is observed (Peixoto et al. 2019). However, understanding the mechanisms of resistance and resilience of corals to environmental change is paramount to ongoing management and restoration efforts at local scales.
The impacts of climate change together with pollution, overexploitation, and habitat destruction severely impact coastal (coral) ecosystems (Hughes et al. 2017). In heavily urbanised regions, physical destruction and eutrophication reduce coral cover and overall diversity (Duprey et al. 2016, 2020) with differential effects on species and individuals. From these, we can identify ‘winners and losers’ associated with any type of environmental change. Those colonies which appear more robust could then be effectively employed for conservation/restoration efforts and utilised to identify mechanisms that contribute to resistance and resilience (Palumbi et al. 2014) and employed for assisted evolution approaches (van Oppen et al. 2015).
Corals are holobionts, relying on an inter-kingdom symbiosis that includes the animal host, its endosymbiotic dinoflagellates from the family Symbiodiniaceae, and a complex suite of other microbial partners including bacteria, archaea, fungi, and viruses (Rosenberg et al. 2007). The members of this holobiont are tightly interlinked and appear to contribute together to its overall health, for instance, through nutrient acquisition and recycling (Bourne et al. 2016). Further, they seem to play an important role in the resilience of their host to environmental change. For example, variation in their associated Symbiodiniaceae can confer thermal resilience (Howells et al. 2012) with shifts in community composition leading to a more advantageous state, at least temporarily (Jones et al. 2008). The coral’s prokaryote community has also been identified as an important contributor to the holobiont’s environmental resilience (Reshef et al. 2006). Changes in the bacterial communities are repeatedly correlated with varying environmental parameters such as temperature or salinity (Röthig et al. 2016; Ziegler et al. 2017).
Water quality can also significantly affect the microbiome of corals (McDevitt-Irwin et al. 2017) and increase the probability of disease outbreaks (Garren et al. 2009; Sutherland et al. 2010). Low water quality and nutrient input, especially in urbanised habitats, result in increases in nitrogen, phosphorus, and turbidity/sedimentation, which affect the local spatial distribution of corals and coral diversity (Duprey et al. 2016). Mostly laboratory-based studies show that increases in nutrients impair function, facilitate disease, and cause bleaching, the latter two often leading to increased chances of mortality (McDevitt-Irwin et al. 2017). However, there is a lack of studies linking specific compounds from multi-stressor eutrophic environments (e.g. ammonium, nitrate, phosphate, or turbidity) directly to shifts in specific coral microbiomes in situ. In most cases, the microbiome shifts or responds to any given stress event prior to visible signs of stress in the coral itself (Bourne et al. 2008). Therefore, it has been suggested that dysbiosis in the coral microbiome could be used as a bioindicator for coral health and contributes to their response to environmental perturbations (Glasl et al. 2019; Sweet et al. 2019).
Corals harbour a relatively high diversity of prokaryotes within their microbiome. It is therefore difficult to untangle key members (belonging to the core microbiome and/or providing important functions) from merely transient associates, for instance, caught in the dynamic mucus layer or simply part of the holobiont’s usual food source (Ainsworth et al. 2015; Sweet and Bulling 2017). Recently, it has been argued that corals, as hosts, may be divided into two main categories as has been previously described for sponges. The first category includes those which regulate a conserved, stable microbiome (‘microbiome regulator’). The second category includes those which have greater ‘microbiome flexibility’ in their associates (‘microbiome conformer’) and therefore the potential for dynamic microbiome adjustment under environmental change (Ziegler et al. 2019). Ziegler et al. (2019) hypothesised that climate change ‘winners’ will be the microbiome conformers which can adjust more readily to environmental change. This extends the coral probiotic hypothesis whereby a coral can counteract a possible negative dysbiosis by adjusting its composition (Reshef et al. 2006).
The above offers a glimmer of hope for the ability of corals to adapt to global change in evolutionarily relatively short timescales. The same mechanisms of microbial turnover may confer intraspecific resilience to local environmental stressors. Hong Kong offers a punctuated water quality gradient that shapes coral distributions on a relatively small spatial scale (Duprey et al. 2016). Mirs Bay in the north-east is flushed by comparatively good quality oceanic waters and harbours at least 94 coral species (Huang et al. 2015). In contrast, to the north-west, heavily eutrophied waters discharging from the Pearl River result in poor water quality, which has driven the regional extirpation of all but one hard coral species, Oulastrea crispata. We capitalised on this species’ ubiquitous distribution to test whether microbial conformity (or regulation) underpins this species’ broad tolerance of environmental conditions. To do this, we examined several water parameters (temperature, salinity, dissolved oxygen (DO), turbidity, pH, ammonia, nitrite, nitrate, total nitrogen, phosphorus, and chlorophyll a) as potential drivers of spatial differences in O. crispata’s microbiome.
Materials and methods
Sampling sites
The marine habitats around Hong Kong are characterised by pronounced environmental fluctuations and intense anthropogenic pressures stemming from the world’s fifth busiest shipping port, coastal development, fishing, and public recreation associated with a city of over seven million inhabitants (Lai et al. 2016). The marine habitats are diverse, with open-ocean conditions to the south-east and estuarine waters in the west and south-west. Seasonality leads to stark environmental changes in Hong Kong waters. In the dry winter season, the north-east monsoon (and the Coriolis force) results in a western current; thus, the effect of Pearl River freshwater inflow on the marine environment of Hong Kong is limited (Lee et al. 2006). In contrast, during the wet summer season, south-western monsoon winds drive the freshwater plume into Hong Kong waters. The Pearl River flows through the most densely populated region in the world and discharges about 350 × 109 m3 yr−1 freshwater, 85 × 106 tons yr−1 sediments, and large amounts of sewage and nutrients (Harrison et al. 2008). The influence of estuarine waters decreases towards the north-east (Mirs Bay, Crescent Island), and this is reflected by a pronounced change in water quality and salinity (Lee et al. 2006). Overall, the water quality is particularly poor in the Pearl River Delta, Deep Bay, and Victoria and Tolo Harbour (Hong Kong Environmental Protection Department 2017).
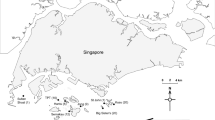
Water quality data from the Hong Kong Environmental Protection Department’s (EPD) fixed sampling stations (1 m depth; Fig. 1) were utilised to characterise the water quality gradient and ultimately identify variables that align with coral microbiome differences. The parameters include temperature, salinity, DO, turbidity, pH (which were all measured with a SEACAT19+ CTD water quality profiler); and ammonia nitrogen, nitrite nitrogen, nitrate nitrogen, total nitrogen, phosphorus, and chlorophyll a concentrations (measured through flow injection analysis) (further details on protocols and the full marine quality database are available at https://www.epd.gov.hk/epd/epic/english/data_marine.html). Nine stations were selected which covered five areas with relatively high coral cover with consideration of local distribution patterns. These included: NM1 for Lantau Island, SM6 and SM18 for Lamma Island, PM9 and PM11 for Bluff Island, MM7 and MM3 for Crescent Island, and TM4 for Centre Island (referred to hereafter as Lantau, Bluff, Lamma, Crescent, and Centre) (Fig. 1). Where two environmental sampling stations approximately equidistant to a field site were available, an average of the two was used. Measurements for this study were taken across three months (June, July, and August) in 2018. The stations outline a pronounced water quality and coral diversity/distribution gradient (Duprey et al. 2016).
Owing to its high latitude location and close proximity to the Pearl River, the environment of Hong Kong is characterised by extremely variable conditions, which contribute to the high local biodiversity (Ng et al. 2017). The region is considered marginal for coral growth (Goodkin et al. 2011). Coral coverage and diversity generally increase with distance from the outflow of freshwater and sediment from the Pearl River Estuary (Goodkin et al. 2011; Duprey et al. 2016). Only one hard coral, Oulastrea crispata (family Faviidae), can cope with the adverse conditions at the sampling site at Lantau. At the opposite side of the water quality gradient, Crescent Island with ~ 60 reported hard coral species (Duprey et al. 2016) provides a site of comparably high coral diversity around Hong Kong. O. crispata is the most widely distributed scleractinian coral in Hong Kong and remarkably robust. Given that O. crispata does not occur closer to the Pearl River Delta, the habitat around Lantau may provide environmental conditions that are at the edge of the coral’s environmental niche. Acclimatisation and/or adaptation to these challenging conditions may therefore include the contribution of all holobiont members. Compared to less challenged individuals, O. crispata specimens from Lantau provide an opportunity to test for a contribution of the microbiome to environmental flexibility.
Coral sample collection and preservation
Five replicate fragments (~ 5 cm2) from the coral Oulastrea crispata were collected at a depth between 3 and 5 m from each of five locations, where possible. At Lantau, Bluff, and Crescent, one replicate each was sampled between 1 and 7 m. To exclude differences in these ‘outliers’, all samples from the same location were tested for consistency in colour, visual health state, and microbiome composition. Coral sampling locations were selected as close to the locations where water parameters were measured as possible, including Lantau (22°19′48.34″ N 114° 1′2.08″ E), Lamma (22°11′27.94″ N 114° 8′8.67″ E), Bluff (22°19′31.98″ N 114°21′15.28″ E), Crescent (22°31′47.75″ N 114°19′0.30″ E), and Centre (22°26′17.10″ N 114°13′20.41″ E) (Fig. 1, Supplementary Table 1). All corals were sampled within two days (18 and 19 August 2018) to reduce temporal variation. All colonies sampled were visually healthy and the same size to reduce any effect of age (Williams et al. 2015). A sterile hammer and chisel were utilised by the divers who wore nitrile gloves. Upon surfacing, the samples were rinsed with filtered seawater, wrapped in sterile aluminium foil, and flash-frozen in liquid nitrogen. All coral fragments were then stored at − 20 °C.
DNA extraction and 16S rRNA gene amplicon sequencing
For all samples, the entire sampled fragment was crushed on liquid nitrogen using bleach-sterilised mortar and pestle. DNA was extracted from the still frozen coral powder using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol with an additional 1-min centrifugation step at 20,000 g after the final washing. DNA concentrations were measured using a Multiskan GO (Thermo Fisher Scientific, Waltham, MA, USA) spectrophotometer. The 16S rRNA amplicons were amplified using the primers 784F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGGATTAGATACCCTGGTA-3′) and 1061R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCRRCACGAGCTGACGAC-3′) (Andersson et al. 2008) with Illumina overhang adaptor sequences (underlined above). The Qiagen multiplex PCR kit was used for amplifications with a total volume of 20 µL containing 10 µL PCR multiplex mix, 0.5 µM of each primer, 1 µL template DNA containing 10–80 ng DNA, and H2O to adjust to the final volume. Amplifications were performed in triplicate for each sample as follows: an initial denaturation at 95 °C for 15 min followed by 27 cycles each with 30 s at 95 °C, 90 s at 55 °C, and 30 s at 72 °C. The final elongation was set at 72 °C for 10 min. PCR amplifications were visually confirmed on a 2% agarose gel using 10 µl PCR products. The triplicate PCR products were pooled for each sample. Library preparation, bead clean-up, pooling in equimolar ratios, and sequencing on a MiSeq Nano v2 PE250 run with 20% PHiX were performed at the Centre for PanorOmic Sciences (CPOS), The University of Hong Kong. Sequences determined in this study have been deposited in the NCBI Sequence Read Archive under accession no. PRJNA609683.
Bacterial community analysis
Bacterial community analysis was conducted using mothur v.1.40.5 (Schloss et al. 2009). Forward and reverse reads were split according to barcodes and assembled to contigs. Contigs < 310 bp and those with ambiguous bases were discarded. Duplicate sequences were merged using the ‘unique.seqs’ command, ‘count.seqs’ was used to keep count of all sequences represented by the unique sequences, and all remaining sequences were aligned against SILVA (Pruesse et al. 2007). Sequences were then quality filtered and pre-clustered, allowing for up to a 2-base pair difference (Huse et al. 2010). Then, singletons, i.e. sequences that occurred only once over all samples, were removed using ‘split.abund’ and chimeric sequences were excluded using VSEARCH as implemented in mothur (Rognes et al. 2016). Remaining sequences were classified against SILVA v.132 using an 80% bootstrap cut-off; then, chloroplasts, mitochondria, archaea, eukaryotes, and unknown sequences were removed. Stacked column plots representing bacterial community compositions at the taxonomic level ‘family’ were constructed. For further analyses, sequences were then clustered into operational taxonomic units (OTUs) with a similarity cut-off of 97%. To account for contamination, OTUs that occurred more than 10% in controls (i.e. sum of extraction and sequencing control) compared to the sum of all other samples were removed (i.e. 132 OTUs removed, 92 occurred only in controls, 40 occurred > 10% in controls, OTU00091 (Pelagibacterium sp.) most abundant contaminant). All samples were then subsampled to 3051 sequences, which corresponds to the lowest number of sequences available for any sample. Alpha diversity indices (Chao 1, Simpson Evenness, and Inverse Simpson Index), principal coordinate analysis (PCoA) (Rao 1964), and linear discriminant analysis effect size (LEfSe) (Segata et al. 2011) were calculated in mothur. To compare significant differences in alpha diversity indices between sites, assumptions were tested, i.e. normal distribution (Shapiro–Wilk W > 0.94, p > 0.1) and homogeneity (pHOMOVA > 0.1), prior to performing single-factor ANOVAs in R (v.3.5.2; R Core Team 2015). The core microbiome was calculated based on subsampled data and includes all OTUs present in all sampled corals (100%).
Correlation and redundancy analyses
Before testing the potential impact of environmental parameters on coral microbiomes, a correlation analysis was performed to test for any interdependencies between environmental parameters. While timescales of coral microbiome response to environmental changes are largely unknown, they may vary between hours and days/weeks (Röthig et al. 2016; Sweet et al. 2017; Ziegler et al. 2017). To account for temporal variations, which, depending on the specific parameter, may have direct, cumulative, and/or lagged effects on the microbiome, three separate analyses were performed. Firstly, all parameters measured in August 2018 (in the month of coral sampling) were tested. Secondly, all parameters, each averaged over July and August 2018, were tested. Thirdly, all parameters, each averaged over June, July, and August 2018, were tested. For each data set, if |R2| value > 0.7, one of the two correlated parameters (whichever correlated with most other parameters) was excluded from further analyses until no correlations were present.
Based on the three data sets with excluded correlated variables, a redundancy analysis was employed to test for and quantify the environmental parameters’ contributions to differences in microbiome community composition between the sites. Further, to avoid the masking of trends, only OTUs with an abundance of 10 or more across all samples were considered (i.e. 1028 OTUs). A permutation test with 999 permutations was employed to test for an overall significance of the redundancy analysis for each of the three environmental data sets. The explanatory power of each parameter was ranked using a stepwise forward and backward selection process that provides rank and statistical significance of each parameter.
Correlation and redundancy analyses, and radar plots were conducted in R using the packages ggcorrplot and vegan.
Results
Environmental data: characterising the water quality gradient
Environmental parameters were measured between June and August 2018 at all eight stations including Lantau, Lamma (two stations), Bluff (two stations), Crescent (two stations), and Centre Islands (Fig. 1). Measured parameters were variable across the five sites (Table 1, Supplementary Table 1). Differences are driven on temporal and spatial scales and reflect the variable nature of the environmental conditions at the survey sites of this study, especially considering that the summer months are characteristically not affected by the regionally strong seasonal variations (Lee et al. 2006). On a spatial scale, averages of the monthly measurements (Table 1) highlight pronounced water quality gradients. Water quality increases from Lantau to Lamma, Bluff, and Crescent, reflected by an average increase in salinity and a decrease in nutrients (ammonia nitrogen, nitrite nitrogen, nitrate nitrogen, and total phosphorus), and chlorophyll a. A second water quality gradient in Tolo Harbour is also present from Centre to Crescent (i.e. an increase in salinity and decrease in ammonia nitrogen, nitrite nitrogen, nitrate nitrogen, and chlorophyll a).
The temporal variability is reflected by pronounced fluctuations at any one site (such as salinity at Lantau, turbidity at Crescent, and total nitrogen at Bluff) (Table 1, Supplementary Table 1). The measurements further include ‘anomalies’ that likely stem from extreme weather events such as strong, seasonal rainfall, e.g. a salinity of 18.2 at Centre and a drop in chlorophyll a from 31.0 to 0.6 μg L−1 within one month at Lantau (Table 1, Supplementary Table 1). These variations will however contribute to the uniqueness of each habitat from which the corals were sampled.
Overall, Lantau was the most variable habitat, as this location exhibits the highest variations for DO, ammonia nitrogen, nitrite nitrogen, total phosphorus, and chlorophyll a, but interestingly the lowest variability for temperature. In contrast, Bluff (most variable for total nitrogen; least variable for DO, turbidity, pH, total phosphorus) together with Crescent (most variable for total nitrogen; least variable for salinity, nitrite nitrogen, nitrate nitrogen, total phosphorus, and chlorophyll a) may be considered more stable during the months assessed in this study.
Correlation analysis: testing for interdependencies between environmental parameters
Each correlation analysis (based on the three environmental data sets: August, AVG July and August, and AVG June, July, and August; Table 1) yielded different correlations (Supplementary Figure 1). After removal of correlating parameters for August 2018, only five out of the initial 11 environmental parameters remained for further analyses (temperature, salinity, DO, nitrite, and total nitrogen). For the averaged months (July and August 2018), four parameters revealed no interdependencies (temperature, salinity, pH, and nitrate). Another four variables (DO, total nitrogen, total phosphorus, and chlorophyll a) were kept for analyses of the averages of June to August 2018.
Bacterial community analysis: characterising the community composition of O. crispata-associated bacterial communities
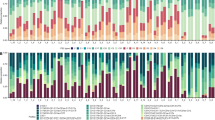
A total of 501,319 sequences were produced across 27 samples (i.e. five Oulastrea crispata samples for each of the five collection sites and two negative controls). After quality trimming and removal of singletons (i.e. sequences that occurred only once across the full data set) and unwanted sequences, 285,955 sequences with an average length of 292 bp were available for subsequent analyses. To evaluate bacterial community composition across samples, we classified all sequences to the family level and visualised the data in a stacked column plot (Fig. 2). Interestingly, no coral microbiome was dominated (in terms of relative abundance) by any one single taxon. The most abundant taxa included an unclassified family of each Gammaproteobacteria, Alphaproteobacteria, Rhodobacteraceae, Flavobacteriaceae, and Vibrionaceae, each contributing less than 10% on average. Variation occurred between coral samples, including those from within the same site. However, the stacked column plots reveal no apparent trend between samples from different sites (Fig. 2). The bacterial community compositions of the coral samples are markedly different from the controls, which were used to identify contaminants (see above) and then disregarded from further analyses.
Bacterial community composition in Oulastrea crispata. Bacterial taxonomy stacked column plot on the phylogenetic level of family (SILVA database, bootstrap ≥ 80). Each colour represents one of the 14 most abundant families with > 6000 sequences each across all samples. All other taxa are grouped under the category ‘others’. Numbers in parentheses depict the number of distinct species within the family. Control 1, extraction control; Control 2, sequencing control
Alpha diversity indices: does O. crispata maintain spatial structural stability in its bacterial associates along the water quality gradient?
To examine differences in the corals’ bacterial community composition (between sites), sequences were clustered into OTUs (similarity cut-off ≤ 0.03). A total of 11,669 distinct OTUs were identified across all five sites and subsampled to 3051 reads per sample. The average number of unique observed OTUs was lowest for the corals collected from Centre at 1173 OTUs and highest for Lamma with 1426 OTUs (Table 2). The average estimated species richness calculated as Chao 1 ranged from 1218 for corals at Centre to 1462 for Lamma. Average Simpson evenness indices were calculated between 0.152 for corals at Bluff and 0.218 for Lantau, while the Inverse Simpson Index had the smallest value of 116 for Bluff and the largest of 198 for Lantau. Upon successful assumptions testing, single-factor ANOVAs were run to test for alpha diversity differences between sites (n = 5 per group, Supplementary Table 2). Despite the pronounced variations in water quality parameters, there were no significant differences (F < 0.5, p > 0.5 for all) in the number of unique OTUs, Chao 1 index, Simpson Evenness Index, or Inverse Simpson Index for any of the corals across the sites surveyed, indicating apparent structural stability in O. crispata’s bacterial associates.
Beta diversity and the core microbiome: characterising spatial differences in O. crispata’s microbiome composition along the water quality gradient
O. crispata is associated with a high diversity of bacterial taxa, which is reflected by 11,669 distinct OTUs across all 25 coral samples (Supplementary Table 3), with an average of 1304 (SD 313) OTUs per sample (Supplementary Table 2). To visualise this high diversity and to identify differences in the corals’ bacterial community profiles, OTU data were plotted in an ordination plot (Fig. 3). The plot shows a separate clustering of samples from Lantau and Bluff and a cluster of four out of five samples from Centre, albeit both axes explaining only ~ 20% of the variation. When statistically assessing differences between the locations, an analysis of molecular variance (AMOVA; 1,000,000 iterations) revealed a significant difference between only two of the sites, Bluff–Lantau (pAMOVA = 0.0049). However, non-significant differences with low p values (pairwise error rate (Bonferroni): 0.005) between Bluff–Centre (pAMOVA = 0.0317), Bluff–Lamma (pAMOVA = 0.03194), Centre and Crescent (pAMOVA = 0.0238), Centre–Lamma (pAMOVA = 0.0239), Centre–Lantau (pAMOVA = p value: 0.0054), Crescent–Lantau (pAMOVA = p value: 0.0053), and Lamma–Lantau (pAMOVA = p value: 0.0080) indicate a generally high spatial heterogeneity. This heterogeneity is further corroborated by the linear discriminant analysis effect size (LEfSe) analysis and the core microbiome (determined as OTUs that occur at 100% of all samples). In total, 286 OTUs were identified as indicator species representing the coral-associated bacterial communities at any given site (LEfSe), including 109 for Lantau, 66 for Lamma, 47 for Bluff, 29 for Crescent, and 35 for Centre (Supplementary Table 4). The core microbiome consisted of seven OTUs (two unclassified Gammaproteobacteria, one unclassified Gammaproteobacteria from the order Incertae Sedis, a Nitrospira sp., an unclassified Sandaracinaceae, an unclassified Cyclobacteriaceae, and a Ruegeria sp.). All core microbiome members are among the 35 most abundant across all samples (> 300 occurrences; Supplementary Table 3). The core microbiome shows a heterogeneous pattern without apparent abundance trends along the environmental gradient or between the sites. That said, one of the identified core members of the microbiome (OTU00014–Cyclobacteriaceae) was also identified as an indicator species (LEfSe) for one of our sites, Lantau. However, its relative abundance at Lantau was only slightly higher than that at both Lamma and Centre (Supplementary Table 3).
Redundancy analysis: linking environmental parameters to coral microbiome variations
To assess the explanatory power of environmental parameters with respect to the response of the coral-associated bacterial communities (i.e. spatial heterogeneity), redundancy analyses were performed on each of the three cleaned environmental data sets (see above ‘Correlation analysis’). The redundancy analysis employed one month of environmental data (closest to the sampling date) and considered temperature, salinity, DO, nitrite, and total nitrogen based on the correlation analysis. It revealed that 22.9% of the observed spatial differences in the microbial communities were explained by the environmental variables included in the model. The results were confirmed by a significant permutation analysis (F4,20 = 1.48, p = 0.001). However, stepwise selection did not show any significance (R2 = 0.022) with total nitrogen explaining most of the microbiome variations (F1 = 1.37, p = 0.146). When considering environmental data from two months (July and August), in this instance including temperature, salinity, pH, and nitrate, similar results were obtained, i.e. 22.9% of the observed variation was explained, further confirmed by a significant permutation analysis (F4,20 = 1.48, p = 0.001). Again, there was no significance in the stepwise selection contributing parameters (R2 = 0.022), with nitrate explaining the most variation (F1 = 1.03, p = 0.458). When three months were taken into account, exploring the impacts of DO, total nitrogen, total phosphorus, and chlorophyll a, 22.9% of the variation was again explained (F4,20 = 1.48, p = 0.001). The stepwise regression was also non-significant (R2 = 0.022) with total phosphorus explaining most of the variation, albeit without statistical significance (F1 = 1.15, p = 0.344). Conserved results in the redundancy analyses irrespective of the considered time frame indicate analytical robustness. However, relatively low explanatory values (i.e. 22.9%) and the lack of significantly contributing environmental parameters suggest other factors play a role.
Discussion
As with many other organisms, coral health depends on the complex yet essential microbiome. However, poor water quality can disrupt the microbiome (dysbiosis) and lead to disease and mortality (Sweet and Bulling 2017). This suggests that microbiome stability itself may contribute to the environmental robustness of the coral holobiont. Indeed, some corals appear to be able to conserve (regulate) their microbiome, while the alternative strategy appears to be microbial conforming (Ziegler et al. 2019). In the latter, corals can adjust their microbial members to gain a temporarily more advantageous composition while maintaining structure and stability. Defining microbiome regulators and conformers has only recently been attempted in corals, and Ziegler et al. (2019) show that common reef-building corals like Pocillopora verrucosa and Acropora hemprichii can utilise these contrasting strategies. P. verrucosa maintained a structurally conserved microbiome along an environmental water quality gradient based on differences in evenness, richness, and diversity. To build on this concept of microbiome flexibility and how the ‘choice’ of strategy might be driven by the environment, we sampled a widely distributed coral, Oulastrea crispata, and assessed its microbiome composition (spatially, not temporally) across an environmental gradient consisting of dramatic changes in water quality. Interestingly, O. crispata’s microbiome appears structurally stable even at the very poor water quality sites.
Coral microbial communities perform essential functions within the holobiont. While the mechanisms are not fully understood, microbes have been linked to holobiont carbon, sulphur, and nitrogen cycling, antimicrobial properties, and secondary metabolism (McDevitt-Irwin et al. 2017). In response to environmental stress, coral microbiome diversity has been shown to increase (Jessen et al. 2013; McDevitt-Irwin et al. 2017), possibly caused by disruption of the native microbiome in terms of stability and function, thereby allowing for invasion of atypical residents (McDevitt-Irwin et al. 2017). The spatial and temporal variability of the microbiome in response to environmental conditions suggests that perhaps only a small number of taxa are obligate symbionts which make up a ‘core microbiome’ (Ainsworth et al. 2015; Hernandez-Agreda et al. 2016; Sweet and Bulling 2017). Poor water quality, in particular when originating from sewage (Wear and Thurber 2015), is known to affect coral physiology and the associated microbiome (Jessen et al. 2013). Studies along eutrophication gradients have demonstrated significant differences in the coral microbiomes according to the water quality/nutrient loads in different species (Ziegler et al. 2016, 2019), and such perturbation characteristically results in dysbiosis and disease (Sutherland et al. 2010). Importantly, upon removal of the stressor, affected corals can quickly reorganise their microbiomes back to their original ‘healthy’ state (Garren et al. 2009; Sweet et al. 2011).
Based on this body of evidence we anticipated, microbiome differences driven by the changes in water parameters would be visible for O. crispata along the west to east, low to high water quality gradient across which this species is distributed in Hong Kong. We show that the microbiome in O. crispata is highly diverse and lacks dominant taxa across a range of environmental conditions (Fig. 2). The coral microbiome at the lowest water quality sites (Lantau and to a lesser extent Centre) differed from sites with relatively better water quality (i.e. Bluff and Crescent), which is characterised by certain indicator species among the rich coral-associated bacterial assemblages (Fig. 3; Supplementary Table 4). Compared to other studies, however, the microbiome differences are moderate. For example, corals exposed to pronounced environmental changes—arguably pushing them to their ‘limit’ in terms of environmental robustness—have been shown to strongly shift or restructure their microbial consortia, either as an acclimation process or as a consequence of dysbiosis. Strongly restructured microbiomes have been observed after exposing corals to 20% increased salinity over 28 days (Röthig et al. 2016), after relocating a deep-sea coral from natural to aquaria settings (Röthig et al. 2017), as a consequence of coral disease (Meyer et al. 2016), and as a result of anthropogenic disturbance (Ziegler et al. 2019). In comparison, O. crispata’s bacterial microbiome exhibited the highest biodiversity in the highest stress site (Lantau), but was structurally stable at all sites based on similar alpha diversity indices (Table 2).
Structural stability in O. crispata exposed to environmental perturbation is further corroborated by distribution patterns of putative pathogens. Environmentally challenged corals often harbour increased numbers of the order Vibrionales that are considered as possible indicators or biomarkers highlighting environmental stress before visual signs of disease are present (Garren et al. 2009; McDevitt-Irwin et al. 2017). However, many other studies have shown such bacteria are present in healthy coral e.g. (McDevitt-Irwin et al. 2017). In this study, Vibrionales are among the most common taxa of the O. crispata microbiome, with Vibrionaceae among the five most abundant families including the most abundant OTU in this study (OTU00001). Interestingly, Vibrionales are more common at sites with better water quality compared to the impacted sites (AVG Lamma 219, Bluff 59, and Crescent 210 vs. Lantau 15 and Centre 57, Supplementary Table 3) and OTU00001 appears an indicator species for a medium-impacted site (Lamma) (Supplementary Table 4). The high contribution of Vibrio spp. found in this study, for all corals, may be the result of all O. crispata showing signs of environmental stress. However, the fact that there was a lower relative abundance of Vibrio spp. in corals from more polluted sites does not support this explanation. Despite all the coral microbiomes being dominated by a high relative number of Vibrio spp., all coral colonies were macroscopically healthy at the time of sampling indicating this corals’ ability to resist a high level of environmental pressure.
Coral reefs typically occur in shallow, oligotrophic, light-flooded tropical regions where environmental parameters are stable (Kleypas et al. 1999) with largely detrimental effects resulting from variation in temperature (Schoepf et al. 2015) and salinity (Coles and Jokiel 1992). Gradients in water quality correlate well with coral diversity and species distribution as shown previously at the same location of this study (Duprey et al. 2016). Examination of the environmental data indicated both high spatial and temporal variations in the measured parameters that create a gradient in water quality along the Hong Kong coastline (Fig. 4, Supplementary Figure 2). Data collected over the three months nearest our sampling times (June to August 2018) included temperatures ranging from 27.3 to 30.4 °C, salinity ranging from 18.2 to 31.8, DO from 4.1 to 8.5 mg L−1, turbidity from 0.6 to 5.3 NTU, pH from 7.7 to 8.5, total nitrogen from 8.21 to 55.69 µmol, and chlorophyll a from 0.6 to 31 μg L−1. These also aligned well with previous notable variations [temperature 13.9 °C (winter)–31.9 °C (summer), salinity 9.8–35.2, pH 7.4–8.5, turbidity 0.3–52 NTU, DO 1.5–11.8 mg L−1, 10.7–678 µmol total nitrogen, 0.2–100 μg L−1 chlorophyll (Hong Kong Environmental Protection Department 2017)].
Radar plots of environmental variables over three months (June, July, and August 2018). Depicted are A means of environmental conditions and B standard deviations for each parameter across the three months. Radial axes for each environmental variable have been scaled so that their ranges are the same for each station. For any particular station, points closer to the centre of the plots represent values that are near the minimum value for the particular environmental variable across all stations and points that are further from the plot centre represent values nearer the maximum value. (Temp, temperature; Sal, salinity; DO, dissolved oxygen; Turb, turbidity; TN, total nitrogen; Phos, phosphate; Chl_a, chlorophyll a)
Despite such variation in the environmental parameters, the corals’ microbiome only partly aligned with the differences. To link microbiome composition and specific water quality parameters, we employed redundancy and regression analyses (see above). Interestingly, the three different correlation analyses performed for this study all indicated slightly different interactions between the parameters, a result which may be driven by the high variability of these parameters across the sites surveyed. Based on the redundancy analyses, the overall contribution of environmental parameters to microbiome differences accounts for only about 23% of the observed variability, and this appears to be largely independent of the considered timescale or parameters. This may be, at least in part, caused by the different distances and time spans for which the environmental parameters and the coral samples were taken (Supplementary Table 1). Surprisingly, we were unable to identify any specific environmental parameter which contributed to the observed microbiome differences. This is in contrast to sediment-associated microbes sampled in the region which showed increased microbial diversity with decreasing water quality (Chen et al. 2019), highlighting the ability of the coral holobiont to structure and maintain a distinct microbial community to that of the surrounding environment (Sweet et al. 2011).
O. crispata’s overall limited microbiome flexibility, based on the spatial structural stability under an extreme environmental gradient, suggests that the coral maintains a conserved microbiome and therefore fits into the ‘microbiome regulator’ category (sensu Ziegler et al. 2019). Given that O. crispata is the most environmentally versatile coral located around Hong Kong, this does not fit with the current working hypothesis that ‘microbiome conformers’ may be more environmentally resilient (Ziegler et al. 2019). However, the previously hypothesised phylogenetic origin of microbiome stability does still correspond with Ziegler et al. (2019). Like P. verrucosa (Ziegler et al. 2019), the ‘microbiome regulator’ O. crispata is (1) from the robust clade, albeit with unresolved evolutionary relationships (Benzoni et al. 2012), and (2) seems to form generally stable associations with its algal endosymbionts (Wong et al. 2016, but see LaJeunesse et al. 2014). The recurring stability of both host–bacterial and host–dinoflagellate associations is an interesting trend that warrants further investigation. Repeated sampling of O. crispata’s microbiome considering Hong Kong’s strong seasonality could gain further insight into its temporal stability and contribution to environmental resilience. Based on the current data, it may be that the wide environmental tolerance of O. crispata is largely a function of the host itself, which is highly flexible in its ecology and physiology. The coral is widely distributed in the Pacific, invasive in the Mediterranean and Korea (Hoeksema and Vicente 2014; Choi and Song 2015), and resistant to both high and low temperatures (Yara et al. 2011). Further, under laboratory conditions, O. crispata reveals a high trophic flexibility and can survive for more than one year in the dark or under daylight conditions in a simple set-up without feeding (Kawaguti 1964; Denis et al. 2012). Finally, O. crispata is a generalist, with a flexible reproduction strategy (asexual and sexual, brooder, and broadcast spawner) (Zayasu et al. 2015). Taken together, these traits point to this coral having a ‘winning’ strategy to survive climate change and other environmental perturbations.
Here, we characterise the bacterial microbiome of Oulastrea crispata along a pronounced water quality gradient in a highly urbanised environment. Our results describe a spatially and temporally highly variable environment along the entire gradient. Reflecting their environment, the coral microbiomes were complex. However, the microbiome variations do not appear pronounced (at least on a spatial scale) and only partially align with environmental differences (explaining ~ 23%). O. crispata appears to be extremely flexible with regard to the environment in which it can thrive. Our data suggest that its bacterial microbiome either contributes little to this ability and/or that microbiome flexibility (i.e. ability to conform) does not necessarily confer environmental robustness to the holobiont. That said, it remains difficult to assign corals into microbiome ‘conformers’ or ‘regulators’ without a direct comparative framework; therefore, more data are required to understand the nature of microbiome flexibility and its potential role in the holobiont’s mechanisms of adaptation to environmental change.
References
Ainsworth T, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M, Gates RD, Padilla-Gamino JL, Spalding HL, Smith C, Woolsey ES, Bourne DG, Bongaerts P, Hoegh-Guldberg O, Leggat W (2015) The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9:2261–2274
Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L (2008) Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3:e2836
Benzoni F, Arrigoni R, Stefani F, Stolarski J (2012) Systematics of the coral genus Craterastrea (Cnidaria, Anthozoa, Scleractinia) and description of a new family through combined morphological and molecular analyses. Syst Biodivers 10:417–433
Bourne D, Iida Y, Uthicke S, Smith-Keune C (2008) Changes in coral-associated microbial communities during a bleaching event. ISME J 2:350–363
Bourne DG, Morrow KM, Webster NS (2016) Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70:317–340
Chen J, McIlroy SE, Archana A, Baker DM, Panagiotou G (2019) A pollution gradient contributes to the taxonomic, functional, and resistome diversity of microbial communities in marine sediments. Microbiome 7:104
Choi E, Song J-I (2015) New Records of Two Zooxanthellate Scleractinian Corals (Anthozoa: Hexacorallia: Scleractinia) from Korea. Anim Syst Evol Divers 31:86–94
Coles SL, Jokiel PL (1992) Effects of salinity on coral reefs. Pollution in Tropical Aquatic Systems. CRC Press, London, pp 147–166
Denis V, Leung JKL, Hsu C-M, Hsieh HJ, Tsai W-S, Chen CA (2012) Dark survival of Oulastrea crispata. Galaxea J Coral Reef Stud 14:117–118
Duprey NN, Wang XT, Kim T, Cybulski JD, Vonhof HB, Crutzen PJ, Haug GH, Sigman DM, Martínez-García A, Baker DM (2020) Megacity development and the demise of coastal coral communities: evidence from coral skeleton δ15N records in the Pearl River estuary. Glob Chang Biol 26:1338–1353
Duprey NN, Yasuhara M, Baker DM (2016) Reefs of tomorrow: eutrophication reduces coral biodiversity in an urbanized seascape. Glob Chang Biol 22:3550–3565
Garren M, Raymundo L, Guest J, Harvell CD, Azam F (2009) Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS ONE 4:e7319
Glasl B, Bourne DG, Frade PR, Thomas T, Schaffelke B, Webster NS (2019) Microbial indicators of environmental perturbations in coral reef ecosystems. Microbiome 7:94
Goodkin NF, Switzer AD, McCorry D, DeVantier L, True JD, Hughen KA, Angeline N, Yang TT (2011) Coral communities of Hong Kong: long-lived corals in a marginal reef environment. Mar Ecol Prog Ser 426:185–196
Harrison PJ, Yin K, Lee JHW, Gan J, Liu H (2008) Physical-biological coupling in the Pearl River Estuary. Cont Shelf Res 28:1405–1415
Hernandez-Agreda A, Leggat W, Bongaerts P, Ainsworth TD (2016) The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. MBio 7:e00560
Hoegh-Guldberg O, Jacob D, Taylor M, Bindi M, Brown S, Camilloni I, Diedhiou A, Djalante R, Ebi K, et al. (2018) Impacts of 1.5 °C Global Warming on Natural and Human Systems. In Global Warming of 1.5 °C; An IPCC Special Report on the impacts of global warming of 1.5 °C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty; Masson-Delmotte V, Zhai P, Pörtner HO, Roberts D, Skea J, Shukla PR, Pirani A, Moufouma-Okia W, Péan C, Pidcock R. et al., Eds.; IPCC: Geneva, Switzerland
Hoeksema BW, Vicente OO (2014) First record of the central Indo-Pacific reef coral Oulastrea crispata in the Mediterranean sea. Mediterr Mar Sci 15:429–436
Hong Kong Environmental Protection Department (2017) Marine water quality in Hong Kong in 2017. https://www.epd.gov.hk/epd/sites/default/files/epd/english/environmentinhk/water/hkwqrc/files/waterquality/annual-report/marinereport2017.pdf
Howells EJ, Beltran VH, Larsen NW, Bay LK, Willis BL, van Oppen MJH (2012) Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat Clim Chang 2:116–120
Huang D, Licuanan WY, Hoeksema BW, Chen CA, Ang PO, Huang H, Lane DJW, Vo ST, Waheed Z, Affendi YA, Yeemin T, Chou LM (2015) Extraordinary diversity of reef corals in the South China Sea. Mar Biodivers 45:157–168
Hughes TP, Barnes ML, Bellwood DR, Cinner JE, Cumming GS, Jackson JBC, Kleypas J, van de Leemput IA, Lough JM, Morrison TH, Palumbi SR, van Nes EH, Scheffer M (2017) Coral reefs in the Anthropocene. Nature 546:82–90
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898
Jessen C, Villa Lizcano JF, Bayer T, Roder C, Aranda M, Wild C, Voolstra CR (2013) In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea Coral Acropora hemprichii. PLoS ONE 8:e62091
Jones AM, Berkelmans R, van Oppen MJ, Mieog JC, Sinclair W (2008) A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc Biol Sci 275:1359–1365
Kawaguti S (1964) An electron microscopic proof for a path of nutritive substances from zooxanthellae to the Reef Coral tissue. Proc Jpn Acad 40:832–835
Kleypas JA, McManus JW, Menez LAB (1999) Environmental limits to coral reef development: Where do we draw the line? Am Zool 39:146–159
Lai RWS, Perkins MJ, Ho KKY, Astudillo JC, Yung MMN, Russell BD, Williams GA, Leung KMY (2016) Hong Kong’s marine environments: History, challenges and opportunities. Reg Stud Mar Sci 8:259–273
LaJeunesse TC, Wham DC, Pettay DT, Parkinson JE, Keshavmurthy S, Chen CA (2014) Ecologically differentiated stress-tolerant endosymbionts in the dinoflagellate genus Symbiodinium (Dinophyceae) Clade D are different species. Phycologia 53:305–319
Lee JHW, Harrison PJ, Kuang C, Yin K (2006) Eutrophication dynamics in Hong Kong coastal waters: Physical and biological interactions. The Environment in Asia Pacific Harbours. Springer, Netherlands, pp 187–206
McDevitt-Irwin JM, Baum JK, Garren M, Vega Thurber RL (2017) Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci 4:262
Meyer JL, Gunasekera SP, Scott RM, Paul VJ, Teplitski M (2016) Microbiome shifts and the inhibition of quorum sensing by Black Band Disease cyanobacteria. ISME J 10:1204–1216
Ng TPT, Cheng MCF, Ho KKY, Lui GCS, Leung KMY, Williams GA (2017) Hong Kong’s rich marine biodiversity: the unseen wealth of South China’s megalopolis. Biodivers Conserv 26:23–36
van Oppen MJH, Oliver JK, Putnam HM, Gates RD (2015) Building coral reef resilience through assisted evolution. Proc Natl Acad Sci 112(8):2307–2313
Palumbi SR, Barshis DJ, Traylor-Knowles N, Bay RA (2014) Mechanisms of reef coral resistance to future climate change. Science 344(6186):895–898
Peixoto R, Sweet M, Bourne D (2019) Customized medicine for corals. Front Mar Sci 6:686
R Core Team (2015) R: A language and environment for statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2012. ISBN 3-900051-07-0
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rao CR (1964) The use and interpretation of principal component analysis in applied research. Sankhyā Indian J Stat Ser A 26:329–358
Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E (2006) The coral probiotic hypothesis. Environ Microbiol 8:2068–2073
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ 4:e2584
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007) The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362
Röthig T, Ochsenkühn MA, Roik A, van der Merwe R, Voolstra CR (2016) Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol Ecol 25:1308–1323
Röthig T, Roik A, Yum LK, Voolstra CR (2017) Distinct bacterial microbiomes associate with the deep-sea coral eguchipsammia fistula from the Red Sea and from Aquaria Settings. Front Mar Sci 4:259
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: Open-Source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schoepf V, Stat M, Falter JL, McCulloch MT (2015) Limits to the thermal tolerance of corals adapted to a highly fluctuating, naturally extreme temperature environment. Sci Rep 5:17639
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Sutherland KP, Porter JW, Turner JW, Thomas BJ, Looney EE, Luna TP, Meyers MK, Futch JC, Lipp EK (2010) Human sewage identified as likely source of white pox disease of the threatened Caribbean elkhorn coral, Acropora palmata. Environ Microbiol 12:1122–1131
Sweet M, Burian A, Fifer J, Bulling M, Elliott D, Raymundo L (2019) Compositional homogeneity in the pathobiome of a new, slow-spreading coral disease. Microbiome 7:139
Sweet MJ, Brown BE, Dunne RP, Singleton I, Bulling M (2017) Evidence for rapid, tide-related shifts in the microbiome of the coral Coelastrea aspera. Coral Reefs 36:1–14
Sweet MJ, Bulling MT (2017) On the importance of the microbiome and pathobiome in coral health and disease. Front Mar Sci 4:9
Sweet MJ, Croquer A, Bythell JC (2011) Dynamics of bacterial community development in the reef coral Acropora muricata following experimental antibiotic treatment. Coral Reefs 30:1121–1133
Wear SL, Thurber RV (2015) Sewage pollution: mitigation is key for coral reef stewardship. Ann N Y Acad Sci 1355(1):15–30
Williams AD, Brown BE, Putchim L, Sweet MJ (2015) Age-related shifts in bacterial diversity in a reef coral. PLoS ONE 10:e0144902
Wong JCY, Thompson P, Xie JY, Qiu JW, Baker DM (2016) Symbiodinium clade C generality among common scleractinian corals in subtropical Hong Kong. Reg Stud Mar Sci 8:439–444
Yara Y, Oshima K, Fujii M, Yamano H, Yamanaka Y, Okada N (2011) Projection and uncertainty of the poleward range expansion of coral habitats in response to sea surface temperature warming: A multiple climate model study. Galaxea J Coral Reef Stud 13:11–20
Zayasu Y, Miyazaki K, Lien Y-T, Okubo N (2015) Direct evidence of sexual reproduction in the zebra coral, Oulastrea crispata (Anthozoa, Scleractinia), in Japan. Invertebr Reprod Dev 59:61–65
Ziegler M, Grupstra CGB, Barreto MM, Eaton M, BaOmar J, Zubier K, Al-Sofyani A, Turki AJ, Ormond R, Voolstra CR (2019) Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun 10:3092
Ziegler M, Roik A, Porter A, Zubier K, Mudarris MS, Ormond R, Voolstra CR (2016) Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Mar Pollut Bull 105:629–640
Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR (2017) Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:14213
Acknowledgements
This study was designed and conducted for the University of Hong Kong ‘Introduction into Molecular Ecology 2018’ workshop; we acknowledge the support of all participants and are thankful for valuable insights on study design from Juan Diego Gaitán-Espitia. A. Burian provided valuable feedback on the manuscript. We further acknowledge helpful feedback from two anonymous reviewers.
Author information
Authors and Affiliations
Contributions
TR, SEM, and DMB designed the study. All authors contributed to data generation and manuscript writing. TR, HB, AC, and TLP analysed and interpreted the data. DMB and MS provided reagents/materials/analysis tools.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Topic Editor Kyle Morgan
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Röthig, T., Bravo, H., Corley, A. et al. Environmental flexibility in Oulastrea crispata in a highly urbanised environment: a microbial perspective. Coral Reefs 39, 649–662 (2020). https://doi.org/10.1007/s00338-020-01938-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-020-01938-2