Abstract
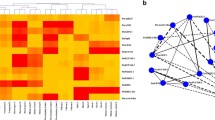
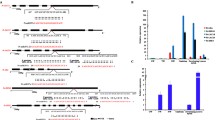
MicroRNAs (miRNAs) play crucial roles in all aspects of plant growth and development, but the genetic interactions of miRNAs and their target genes in woody plants are largely unknown. Here, we integrated association genetics and expression profiling to decipher the allelic variations and interactions of the Pto-MIR319 family of miRNAs and 12 putative Pto-miR319 target genes related to wood formation in 435 unrelated individuals of Populus tomentosa Carrière (Chinese white poplar). Expression pattern analysis showed that among all pairings between expressions of pre-miRNA of Pto-MIR319 members and targets, 70.0% showed negative correlation of expression levels (r = – 0.944 to 0.674, P < 0.01) in eight tissues and organs of poplar, suggesting that Pto-miR319 may participate in the regulatory network of wood formation. Single SNP-based association studies identified 137 significant associations (P < 0.01, Q < 0.1), representing 126 unique SNPs from Pto-MIR319 members and their targets, with 10 tree growth traits, revealing that these genetic factors have common roles related to wood formation. Epistasis analysis uncovered 105 significant SNP–SNP associations (P < 0.01) influencing the 10 traits, demonstrating the close genetic interactions between Pto-MIR319 family members and the 12 Pto-miR319 target genes. Notably, one common SNP, in the precursor region of Pto-MIR319e, affected the stability of Pto-MIR319e’s secondary structure by altering the stem-loop structure and minimum free energy, contributing to variations in the expression of Pto-MIR319e and Pto-miR319e target genes. This study enriches the understanding of the functions of miR319 family miRNAs in poplar and exemplifies a feasible approach to exploring the genetic effects underlying miRNA–mRNA interactions related to complex traits in trees.




Similar content being viewed by others
Data availability
Sequence data in this article have been deposited with the NCBI GenBank Data Library under the accession codes listed in Table S2, and the degradome sequencing data are available in the Genome Sequence Archive of the BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Science, under the accession code CRA000989.
References
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18(10):758–762. https://doi.org/10.1016/j.cub.2008.04.042
Aggarwal P, Das Gupta M, Joseph AP, Chatterjee N, Srinivasan N, Nath U (2010) Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22(4):1174–1189. https://doi.org/10.1105/tpc.109.066647
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Beaulieu J, Doerksen T, Boyle B, Clement S, Deslauriers M, Beauseigle S et al (2011) Association genetics of wood physical traits in the conifer white spruce and relationships with gene expression. Genetics 188(1):197–214. https://doi.org/10.1534/genetics.110.125781
Bensen JT, Tse CK, Nyante SJ, Barnholtz-Sloan JS, Cole SR, Millikan RC (2013) Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: the Carolina Breast Cancer Study. Cancer Cause Control 24(6):1099–1109. https://doi.org/10.1007/s10552-013-0187-z
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579(26):5923–5931. https://doi.org/10.1016/j.febslet.2005.07.071
Chen J, Qin Z, Jiang Y, Wang Y, He Y, Dai J et al (2014) Genetic variations in the flanking regions of miR-101-2 are associated with increased risk of breast cancer. PLoS ONE 9(1):e86319. https://doi.org/10.1371/journal.pone.0086319
Chen B, Chen J, Du Q et al (2018) Genetic variants in micro RNA biogenesis genes as novel indicators for secondary growth in Populus. New Phytol 219(4):1263–1282. https://doi.org/10.1111/nph.15262
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–159. https://doi.org/10.1093/nar/gkr319
Dmitriev AA, Kudryavtseva AV, Bolsheva NL, Zyablitsin AV, Rozhmina TA, Kishlyan NV et al (2017) miR319, miR390, and miR393 are involved in aluminum response in Flax (Linum usitatissimum L.). Biomed Res Int. https://doi.org/10.1155/2017/4975146
Du Q, Wang B, Wei Z, Zhang D, Li B (2012) Genetic diversity and population structure of Chinese white poplar (Populus tomentosa) revealed by SSR markers. J Hered 103(6):853–862. https://doi.org/10.1093/jhered/ess061
Du Q, Xu B, Pan W, Gong C, Wang Q, Tian J et al (2013) Allelic variation in a cellulose synthase gene (PtoCesA4) associated with growth and wood properties in Populus tomentosa. Genes Genom Genet 3(11):2069–2084. https://doi.org/10.1534/g3.113.007724
Du Q, Xu B, Gong C, Yang X, Pan W, Tian J (2014) Variation in growth, leaf, and wood property traits of Chinese white poplar (Populus tomentosa), a major industrial tree species in northern China. Can J Forest Res 44:326–339
Du Q, Tian J, Yang X, Pan W, Xu B, Li B et al (2015) Identification of additive, dominant, and epistatic variation conferred by key genes in cellulose biosynthesis pathway in Populus tomentosa. DNA Res 22(1):53–67. https://doi.org/10.1093/dnares/dsu040
Duan R, Pak C, Jin P (2007) Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 16(9):1124–1131. https://doi.org/10.1093/hmg/ddm062
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Gong J, Tong Y, Zhang HM, Wang K, Hu T, Shan G et al (2012) Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat 33(1):254–263. https://doi.org/10.1002/humu.21641
Gu M, Xu K, Chen A, Zhu Y, Tang G, Xu G (2010) Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol Plant 138(2):226–237. https://doi.org/10.1111/j.1399-3054.2009.01320.x
Hahn LW, Ritchie MD, Moore JH (2003) Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19(3):376–382
Hardy O, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. https://doi.org/10.1046/j.1471-8278
Hu T, Chen YZ, Kiralis JW, Collins RL, Wejse C, Sirugo G et al (2013) An information-gain approach to detecting three-way epistatic interactions in genetic association studies. J Am Med Inform Assn 20(4):630–636. https://doi.org/10.1136/amiajnl-2012-001525
Ingvarsson PK (2008) Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180(1):329–340. https://doi.org/10.1534/genetics.108.090431
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Bio 57:19–53. https://doi.org/10.1146/annurev.arplant.57.032905.105218
Kall L, Storey JD, MacCoss MJ, Noble WS (2008) Posterior error probabilities and false discovery rates: two sides of the same coin. J Proteome Res 7(1):40–44. https://doi.org/10.1021/pr700739d
Karakülah G, Yücebilgili KK, Unver T (2016) PeTMbase: a database of plant endogenous target mimics (eTMs). PLoS ONE 11(12):e167698. https://doi.org/10.1371/journal.pone.0167698
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(Database issue):D68–73. https://doi.org/10.1093/nar/gkt1181
Li X, Wu HX, Dillon SK, Southerton SG (2009) Generation and analysis of expressed sequence tags from six developing xylem libraries in Pinus radiata D. Don BMC Genomics 10:41. https://doi.org/10.1186/1471-2164-10-41
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Lu S, Li Q, Wei H, Chang MJ, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun YH, Yuan L, Yeh TF, Peszlen I, Ralph J, Sederoff RR, Chiang VL (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA 110(26):10848–10853. https://doi.org/10.1073/pnas.1308936110
Mackay TFC (2014) Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet 15(1):22–33. https://doi.org/10.1038/nrg3627
Martin-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15(1):31–39. https://doi.org/10.1016/j.tplants.2009.11.003
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL et al (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20(12):3186–3190. https://doi.org/10.1105/tpc.108.064311
Neale DB (2007) Genomics to tree breeding and forest health. Curr Opin Genet Dev 17(6):539–544. https://doi.org/10.1016/j.gde.2007.10.002
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12(2):111–122. https://doi.org/10.1038/nrg2931
Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R et al (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell 13(1):115–125. https://doi.org/10.1016/j.devcel.2007.04.012
Porth I, Klapste J, Skyba O, Lai BS, Geraldes A, Muchero W et al (2013) Populus trichocarpa cell wall chemistry and ultrastructure trait variation, genetic control and genetic correlations. New Phytol 197(3):777–790. https://doi.org/10.1111/nph.12014
Quan M, Wang Q, Phangthavong S, Yang X, Song Y, Du Q et al (2016) Association studies in Populus tomentosa reveal the genetic interactions of Pto-MIR156c and its targets in wood formation. Front Plant Sci 7:1159. https://doi.org/10.3389/fpls.2016.01159
Rawlings-Goss RA, Campbell MC, Tishkoff SA (2014) Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med Genomics 7:53. https://doi.org/10.1186/1755-8794-7-53
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J et al (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98(20):11479–11484. https://doi.org/10.1073/pnas.201394398
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25(7):2383–2399. https://doi.org/10.1105/tpc.113.113159
Ryan BM, Robles AI, Harris CC (2010) Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 10(6):389–402. https://doi.org/10.1038/nrc2867
Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE et al (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6(9):e230. https://doi.org/10.1371/journal.pbio.0060230
Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF (2014) Repression of cell proliferation by miR319-regulated TCP4. Mol Plant 7(10):1533–1544. https://doi.org/10.1093/mp/ssu084
Shamimuzzaman M, Vodkin L (2012) Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics 13:310. https://doi.org/10.1186/1471-2164-13-310
Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA et al (2009) SNPs in human miRNA genes affect biogenesis and function. RNA 15(9):1640–1651. https://doi.org/10.1261/rna.1560209
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Wang ST, Sun XL, Hoshino Y, Yu Y, Jia B, Sun ZW et al (2014) MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 9(3):e91357. https://doi.org/10.1371/journal.pone.0091357
Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122(4):1003–1013
Xiao L, Quan MY, Du QZ, Chen JH, Xie JB, Zhang DQ (2017) Allelic interactions among Pto-MIR475b and its four target genes potentially affect growth and wood properties in Populus. Front Plant Sci 8:1055. https://doi.org/10.3389/Fpls.2017.01055
Xie M, Zhang S, Yu B (2015) microRNA biogenesis, degradation and activity in plants. Cell Mol Life Sci 72(1):87–99. https://doi.org/10.1007/s00018-014-1728-7
Xie J, Yang X, Song Y, Du Q, Li Y, Chen J et al (2017) Adaptive evolution and functional innovation of Populus-specific recently evolved microRNAs. New Phytol 213(1):206–219. https://doi.org/10.1111/nph.14046
Yoo SS, Hong MJ, Lee JH et al (2017) Association between polymorphisms in micro RNA target sites and survival in early-stage non-small cell lung cancer. Thorac Cancer 8(6):682–686. https://doi.org/10.1111/1759-7714.12478
Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG (2002) Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 53(2):79–91. https://doi.org/10.1159/000057986
Zhang BH, Wang QL, Pan XP (2007) MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210(2):279–289. https://doi.org/10.1002/jcp.20869
Zhang D, Du Q, Xu B, Zhang Z, Li B (2010) The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol Genet Genomics 284(2):105–119. https://doi.org/10.1007/s00438-010-0552-5
Zhang L, Liu Y, Song F, Zheng H, Hu L, Lu H et al (2011) Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci USA 108(33):13653–13658. https://doi.org/10.1073/pnas.1103360108
Zhang B, Unver T (2018) A critical and speculative review on microrna technology in crop improvement: current challenges and future directions. Plant Sci 274:193–200. https://doi.org/10.1016/j.plantsci.2018.05.031
Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X et al (2015) Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot 66(15):4653–4667. https://doi.org/10.1093/jxb/erv238
Acknowledgements
This work was supported by the Projects of the National Natural Science Foundation of China (Nos. 31670333 and 31401138) and the China Postdoctoral Science Foundation (Nos. 2018M640084 and 2019T120058).
Author information
Authors and Affiliations
Contributions
DZ designed the research; JS, MQ, and LX performed the experiments; JX and QD collected and analyzed the data; JS and MQ wrote the manuscript; DZ obtained funding and is responsible for this article. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2020_1667_MOESM2_ESM.tif
Figure S2. Transcript abundances of Pto-MIR319e and its five target genes under different genotypes of Pto-MIR319e_SNP5. Error bars indicate the SE of biological replicates
438_2020_1667_MOESM8_ESM.docx
Table S6. The common single-nucleotide polymorphisms (SNPs; minor allele frequency ≥ 5%) in Pto-MIR319a–h and their 12 target genes
438_2020_1667_MOESM9_ESM.docx
Table S7. Detail of significant SNPs within the Pto-MIR319 family and their targets associated with tree growth and wood formation in a natural population of P. tomentosa.
438_2020_1667_MOESM10_ESM.docx
Table S8. Significant haplotypes from Pto-MIR319 family members and the target genes associated with growth and wood properties in the P. tomentosa association population.
438_2020_1667_MOESM11_ESM.docx
Table S9. The SNP pairs and their main effects detected from Pto-MIR319 family members and the 12 targets under the epistasis model in the P. tomentosa association population.
Rights and permissions
About this article
Cite this article
Si, J., Quan, M., Xiao, L. et al. Genetic interactions among Pto-miR319 family members and their targets influence growth and wood properties in Populus tomentosa. Mol Genet Genomics 295, 855–870 (2020). https://doi.org/10.1007/s00438-020-01667-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01667-9