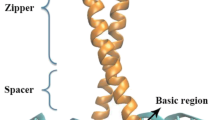
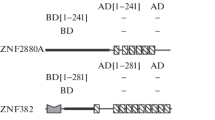
Abstract—The large amount of the DNA- and RNA-binding protein regulatory factors contain Zn-domains. Total six Zn-CysHis factor families with different functions were considered. Among them the transcription factors of family PF00096, complex type Zn-Cys2His2, is the most important because it includes over 340.000 members of 365.000 total members listed at present in the Zn-containing factor families. The role of electrostatic potential of Zn-domains in the structure and function of factors from different factor families has been considered. We have shown that the positive electrostatic potential of Zn-domains plays an essential role in forming contact of factors’ Zn-domains with DNA/RNA in three protein factor families PF00096, PF12874, and PF09329. While in other three families PF03119, PF08996, and PF01258 Zn-domains do not contact with nucleic acids, and electrostatic interactions do not play a distinctive role, consequently.



Similar content being viewed by others
REFERENCES
Klug A. 2010. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Ann. Rev. Biochem.79, 213–231.
Polozov R.V., Sivozhelezov V.S., Chirgadze Y.N., Ivanov V.V. 2015. Recognition rules for binding of Zn-Cys2His2 transcription factors to operator DNA. J. Biomol. Struct. Dyn. 33, 253‒266.
Chirgadze Y.N., Boshkova E.A., Polozov R.V., Sivozhelezov V.S., Dzyabchenko A.V., Kuzminsky M.B., Stepanenko V.A., Ivanov V.V. 2018. The electrostatic role of the Zn-Cys2His2 complex in binding of operator DNA with transcription factors: Mouse EGR-1 from the Cys2His2 family. J. Biomol. Struct. Dyn. 36, 3902‒3915.
Engelke D.R., Ng S.Y., Shastry B.S., Roeder RG. 1980. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 19, 717‒728.
Miller J., McLachlan A.D., Klug A. 1985. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes.EMBO J.4, 1609‒1614.
Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E.L., Eddy S.R., Bateman A., Finn R.D. 2012. The Pfam protein families’ database. Nucleic Acids Res.40, D290–D301.
Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. 2013. GenBank. Nucleic Acids Res.41, 36‒42.
Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., et al. 2002. The protein data bank. Acta Cryst. 58, Pt.6 (1), 899‒907.
Wuttke D.S., Foster M.P., Case D.A., Gottesfeld J.M., Wright P.E. 1997. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: Determinants of affinity and sequence specificity. J. Mol. Biol.273, 183‒206.
Elrod-Erickson M., Rould M.A., Nekludova L., Pabo C.O. 1996. Zif268 protein–DNA complexes refined at 1.6 Å: A model system for understanding zinc finger–DNA interactions. Structure. 4, 1171‒1180.
DeLano W.L., Ultsch M.H., de Vos A.M., Wells J.A. 2000. Convergent solutions to binding at a protein–protein interface. Science.287, 1279‒1283.
Nolte R.T., Conlin R.M., Harrison S.C., Brown R.S. 1998. Differing roles for zinc fingers in DNA recognition: Structure of a six-finger transcription factor IIIA complex. Proc. Natl. Acad. Sci. U. S. A.95, 2938‒2943.
Chirgadze Y.N., Boshkova E.A., Yakovlev A.V., Ivanov V.V. 2019. Side projections of double-helical DNA: Example of binding patterns of DNA in the complex with factor TFIIIA. J. Biomol. Struct. Dyn.37, 4433‒4436.
Burge R.G., Martinez-Yamout M.A., Dyson H.J., Wright P.E. 2014. Structural characterization of interactions between the double-stranded RNA-binding zinc finger protein JAZ and nucleic acids. Biochemistry.53, 1495‒1510.
Warren E.M., Huang H., Fanning E., Chazin W.J., Eichman B.F. 2009. Physical interactions between Mcm10, DNA, and DNA polymerase α. J. Biol. Chem. 284, 24662‒24672.
Nandakumar J., Nair P.A., Shuman S. 2007. Last stop on the road to repair: Structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol. Cell.26, 257‒271.
Suwa Y., Gu J., Baranovskiy A.G., Babayeva N.D., Pavlov Y.I., Tahirov T.H. 2015. Crystal structure of the human Pol αB subunit in complex with the C-terminal domain of the catalytic subunit domain of the catalytic subunit. J. Biol. Chem. 290, 14328‒14337.
Perederina A., Svetlov V., Vassylyeva M.N., Tahirov T.H., Yokoyama S., Artsimovitch I., Vassylyev D.G. 2004. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell. 118, 297‒309.
ACKNOWLEDGMENTS
Authors would like to thank Ms Eugenia A. Boshkova, Institute of Protein Research Russian Academy of Sciences, for the help in working with databases and useful discussions.
Funding
The research was supported by the Russian Foundation for Basic Research (project nos. 17-07-01331a and 14-03-01091a).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. No potential conflict of interest was reported by the authors.
Rights and permissions
About this article
Cite this article
Chirgadze, Y.N., Ivanov, V.V. Zn-CysHis Protein Factor Families: Role of Electrostatic Interaction of Zn-Domains in Factor Functions. Mol Biol 54, 157–162 (2020). https://doi.org/10.1134/S002689332002003X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002689332002003X