Abstract
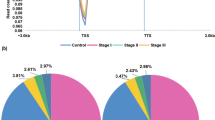
DNA hypermethylation and mutations are key mechanisms for the downregulation of tumor suppressor genes. NotI-microarrays allowed us to detect hypermethylation and/or deletions in 180 NotI sites associated with 188 genes of human chromosome 3, in 24 paired (tumor/normal) colon samples. The most frequent aberrations (in more than 20% of tumor samples) were detected in the promoter regions of 20 genes. Expression and promoter methylation of these genes were analyzed using the data for paired colon samples from The Cancer Genome Atlas project. Three genes—ALDH1L1, PLCL2, and PPP2R3A—revealed a more than two-fold average decrease in expression and a negative correlation between mRNA level and promoter hypermethylation. The expression of these three genes was then evaluated in 30 paired colon samples by quantitative PCR. Frequent (in more than 60% of cases) and significant (5–9-fold on average) mRNA level decrease was found for each of the genes in the tumor samples. The results indicate a suppressor role of the ALDH1L1, PLCL2, and PPP2R3A genes in colon cancer, as well as functional significance of hypermethylation in the downregulation of these genes.


Similar content being viewed by others
REFERENCES
Kuipers E.J., Grady W.M., Lieberman D., Seufferlein T., Sung J.J., Boelens P.G., van de Velde C.J., Watanabe T. 2015. Colorectal cancer. Nat. Rev. Dis. Primers. 1, 15065.
The Cancer Genome Atlas Network. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487, 330‒337.
Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. 2013. Cancer genome landscapes. Science. 339, 1546‒1558.
Alexander J., Watanabe T., Wu T.T., Rashid A., Li S., Hamilton S.R. 2001. Histopathological identification of colon cancer with microsatellite instability. Am. J. Pathol. 158, 527‒535.
Fearon E.R. 2011. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 6, 479‒507.
Kudryavtseva A.V., Lipatova A.V., Zaretsky A.R., Moskalev A.A., Fedorova M.S., Rasskazova A.S., Shibukhova G.A., Snezhkina A.V., Kaprin A.D., Alekseev B.Y., Dmitriev A.A., Krasnov G.S. 2016. Important molecular genetic markers of colorectal cancer. Oncotarget. 7, 53959‒53983.
Webber C., Gospodarowicz M., Sobin L.H., Wittekind C., Greene F.L., Mason M.D., Compton C., Brierley J., Groome P.A. 2014. Improving the TNM classification: Findings from a 10-year continuous literature review. Int. J. Cancer. 135, 371‒378.
Li J., Protopopov A., Wang F., Senchenko V., Petushkov V., Vorontsova O., Petrenko L., Zabarovska V., Muravenko O., Braga E., Kisselev L., Lerman M.I., Kashuba V., Klein G., Ernberg I., et al. 2002. NotI subtraction and NotI-specific microarrays to detect copy number and methylation changes in whole genomes. Proc. Natl. Acad. Sci. U. S. A.99, 10724‒10729.
Dmitriev A.A., Kashuba V.I., Haraldson K., Senchenko V.N., Pavlova T.V., Kudryavtseva A.V., Anedchenko E.A., Krasnov G.S., Pronina I.V., Logi-nov V.I., Kondratieva T.T., Kazubskaya T.P., Braga E.A., Yenamandra S.P., Ignatjev I., et al. 2012. Genetic and epigenetic analysis of non-small cell lung cancer with NotI-microarrays. Epigenetics. 7, 502‒513.
Krasnov G.S., Kudryavtseva A.V., Snezhkina A.V., Lakunina V.A., Beniaminov A.D., Melnikova N.V., Dmitriev A.A. 2019. Pan-cancer analysis of TCGA data revealed promising reference genes for qPCR normalization. Front. Genet. 10, 97.
Krasnov G.S., Oparina N.Yu., Dmitriev A.A., Kudryavtseva A.V., Anedchenko E.A., Kondrat’eva T.T., Zabarovskii E.R., Senchenko V.N. 2011. RPN1, a new reference gene for quantitative data normalization in lung and kidney cancer. Mol. Biol. (Moscow). 45 (2), 211‒220.
Melnikova N.V., Dmitriev A.A., Belenikin M.S., Koroban N.V., Speranskaya A.S., Krinitsina A.A., Krasnov G.S., Lakunina V.A., Snezhkina A.V., Sadritdinova A.F., Kishlyan N.V., Rozhmina T.A., Klimina K.M., Amosova A.V., Zelenin A.V., et al. 2016. Identification, expression analysis, and target prediction of flax genotroph microRNAs under normal and nutrient stress conditions. Front. Plant Sci. 7, 399.
Senchenko V.N., Kisseljova N.P., Ivanova T.A., Dmitriev A.A., Krasnov G.S., Kudryavtseva A.V., Panasenko G.V., Tsitrin E.B., Lerman M.I., Kisseljov F.L., Kashuba V.I., Zabarovsky E.R. 2013. Novel tumor suppressor candidates on chromosome 3 revealed by NotI-microarrays in cervical cancer. Epigenetics. 8, 409‒420.
Haraldson K., Kashuba V.I., Dmitriev A.A., Senchenko V.N., Kudryavtseva A.V., Pavlova T.V., Braga E.A., Pronina I.V., Kondratov A.G., Rynditch A.V., Lerman M.I., Zabarovsky E.R. 2012. LRRC3B gene is frequently epigenetically inactivated in several epithelial malignancies and inhibits cell growth and replication. Biochimie. 94, 1151‒1157.
Glantz S.A. 2005. Primer of Biostatistics. New York, USA: McGraw-Hill.
Krasnov G.S., Dmitriev A.A., Melnikova N.V., Zaretsky A.R., Nasedkina T.V., Zasedatelev A.S., Senchenko V.N., Kudryavtseva A.V. 2016. CrossHub: A tool for multi-way analysis of The Cancer Genome Atlas (TCGA) in the context of gene expression regulation mechanisms. Nucleic Acids Res.44, e62.
Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P. 1999. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. U. S. A.96, 8681‒8686.
Ghosh A., Ghosh S., Maiti G.P., Sabbir M.G., Zabarovsky E.R., Roy A., Roychoudhury S., Panda C.K. 2010. Frequent alterations of the candidate genes hMLH1, ITGA9 and RBSP3 in early dysplastic lesions of head and neck: Clinical and prognostic significance. Cancer Sci.101, 1511‒1520.
Beniaminov A.D., Krasnov G.S., Dmitriev A.A., Puzanov G.A., Snopok B.A., Senchenko V.N., Kashuba V.I. 2016. Interaction of two tumor suppressors: phosphatase CTDSPL and Rb protein. Mol. Biol. (Moscow). 50 (3), 438–441.
Krupenko S.A., Krupenko N.I. 2018. ALDH1L1 and ALDH1L2 folate regulatory enzymes in cancer. Adv. Exp. Med. Biol. 1032, 127‒143.
Beniaminov A.D., Puzanov G.A., Krasnov G.S., Kaluzhny D.N., Kazubskaya T.P., Braga E.A., Kudryavtseva A.V., Melnikova N.V., Dmitriev A.A. 2018. Deep sequencing revealed a CpG methylation pattern associated with ALDH1L1 suppression in breast cancer. Front. Genet.9, 169.
Oleinik N.V., Krupenko N.I., Krupenko S.A. 2011. Epigenetic silencing of ALDH1L1, a metabolic regulator of cellular proliferation, in cancers. Genes Cancer. 2, 130‒139.
Dmitriev A.A., Rudenko E.E., Kudryavtseva A.V., Krasnov G.S., Gordiyuk V.V., Melnikova N.V., Stakhovsky E.O., Kononenko O.A., Pavlova L.S., Kondratieva T.T., Alekseev B.Y., Braga E.A., Senchenko V.N., Kashuba V.I. 2014. Epigenetic alterations of chromosome 3 revealed by NotI-microarrays in clear cell renal cell carcinoma. BioMed. Res. Internat.2014, 735292.
Mazhar S., Taylor S.E., Sangodkar J., Narla G. 2019. Targeting PP2A in cancer: Combination therapies. Biochim. Biophys. Acta.Mol. Cell Res. 1866, 51‒63.
ACKNOWLEDGMENTS
Assessment of gene expression by the quantitative PCR was performed using the equipment of EIMB RAS “Genome” center (http://www.eimb.ru/ru1/ckp/ccu_genome_c.php).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
FUNDING
This work was financially supported by the Russian Science Foundation (grant 17-74-20064).
COMPLIANCE WITH ETHICAL STANDARDS
All procedures performed in this work comply with the ethical standards of the institutional committee for research ethics and the 1964 Helsinki Declaration and its subsequent changes, or comparable ethical standards.
Conflict of interests. The authors declare no conflict of interest.
Additional information
Translated by P. Vikhreva
Abbreviations: CC, colon cancer; CIN, chromosome instability; MSI, microsatellite instability; CIMP, CpG island methylator phenotype.
Rights and permissions
About this article
Cite this article
Dmitriev, A.A., Beniaminov, A.D., Melnikova, N.V. et al. Functional Hypermethylation of ALDH1L1, PLCL2, and PPP2R3A in Colon Cancer. Mol Biol 54, 178–184 (2020). https://doi.org/10.1134/S0026893320010057
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893320010057