Abstract
Purpose
Cryptosporidium parvum is an Apicomplexa parasite that causes watery diarrhea (cryptosporidiosis), especially in children and immunocompromised adults (the latter in a very severe form). No effective treatment exists against infection by this parasite. Phosphatases participate in the regulation of various cellular functions and are thus considered potential therapeutic targets in many diseases. The aim of the present study was to indirectly identify and in silico characterize a protein phosphatase 2C of C. parvum.
Methods
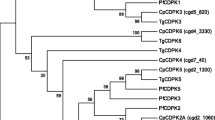
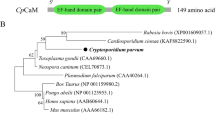
Western blot and indirect immunofluorescence microscopy were performed with a polyclonal antibody against Leishmania major PP2C. Possible cross-reactivity with LmPP2C was assessed by in silico sequence homology to analyze phylogenetic relationships between distinct C. parvum PP2Cs. In addition, another bioinformatics approach was used to predict the possible relationship and function of C. parvum PP2C in the regulation of several cellular processes.
Results
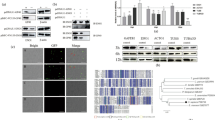
Western blotting showed a protein of approximately 72 kDa. With immunofluorescence, PP2C was localized in the nucleus of oocysts (with some additional labeling in the cytoplasm) and at the apical region of sporozoites. By aligning C. parvum PP2C with known ortholog sequences and carrying out PPI analysis, a determination could be made of the degree of conservation of these enzymes, their possible relationship, and their function in the regulation of several cellular processes associated with a likely nuclear location.
Conclusion
Microscopic localization by immunofluorescence identified CpPP2C at the nucleus in oocysts and at the apical end of the sporozoite body. Hence, this enzyme could be associated with proteins that have an important role in the regulation of transcription and other processes orchestrated by MAPK kinases, according to in silico analysis.





Similar content being viewed by others
References
Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA et al (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304(5669):441–445. https://doi.org/10.1126/science.1094786
Aguirre-García MM, Okhuysen PC (2007) Cryptosporidium parvum: identification and characterization of an acid phosphatase. Parasitol Res 101(1):85–89. https://doi.org/10.1007/s00436-006-0457-8
Andreeva AV, Kutuzov MA (2004) Widespread presence of "bacterial-like" PPP phosphatases in eukaryotes. BMC Evol Biol 4:47. https://doi.org/10.1186/1471-2148-4-47
Arrowood MJ, Sterling CR (1987) Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J Parasitol 73(2):314–319
Artz JD, Wernimont AK, Allali-Hassani A, Zhao Y, Amani M, Lin Y-H et al (2011) The Cryptosporidium parvum kinome. BMC Genom 12(1):478. https://doi.org/10.1186/1471-2164-12-478
Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M et al (2009) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38(suppl_1):D457–D462. https://doi.org/10.1093/nar/gkp851
Bones AJ, Jossé L, More C, Miller CN, Michaelis M, Tsaousis AD (2019) Past and future trends of Cryptosporidium in vitro research. Exp Parasitol 196:28–37. https://doi.org/10.1016/j.exppara.2018.12.001
Bork P, Brown NP, Hegyi H, Schultz J (1996) The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci 5(7):1421–1425. https://doi.org/10.1002/pro.5560050720
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Brumlik MJ, Pandeswara S, Ludwig SM, Murthy K, Curiel TJ (2011) Parasite mitogen-activated protein kinases as drug discovery targets to treat human protozoan pathogens. J Signal Transduct. https://doi.org/10.1155/2011/971968
Burns JM Jr, Parsons M, Rosman DE, Reed SG (1993) Molecular cloning and characterization of a 42-kDa protein phosphatase of Leishmania chagasi. J Biol Chem 268:17155–17161
Cohen P (2002) The origins of protein phosphorylation. Nat Cell Biol 4:E127–130. https://doi.org/10.1038/ncb0502-e127
Das AK, Helps NR, Cohen PT, Barford D (1996) Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J 15:6798–6809
Dorin-Semblat D, Quashie N, Halbert J, Sicard A, Doerig C, Peat E et al (2007) Functional characterization of both MAP kinases of the human malaria parasite Plasmodium falciparum by reverse genetics. Mol Microbiol 65(5):1170–1180. https://doi.org/10.1111/j.1365-2958.2007.05859.x
DuPont HL, Chappell CL, Sterling CR, Okhuysen PC, Rose JB, Jakubowski W (1995) The infectivity of Cryptosporidium parvum in healthy volunteers. N Engl J Med 332(13):855–859. https://doi.org/10.1056/NEJM199503303321304
Escalona-Montaño AR, Pérez-Montfort R, Cabrera N, Mondragón-Flores R, Vélez-Ramírez DE, Gómez-Sandoval JN et al (2017) Protein phosphatase PP2C in the flagellum of Leishmania major: cloning and characterization. Parasitol Open. https://doi.org/10.1017/pao.2017.14
Fuchs S, Grill E, Meskiene I, Schweighofer A (2013) Type 2C protein phosphatases in plants. FEBS J 280(2):681–693. https://doi.org/10.1111/j.1742-4658.2012.08670.x
Gomez de Leon CT, Diaz Martin RD, Mendoza Hernandez G, Gonzalez Pozos S, Ambrosio JR, Mondragon Flores R (2014) Proteomic characterization of the subpellicular cytoskeleton of Toxoplasma gondii tachyzoites. J Proteom 111:86–99. https://doi.org/10.1016/j.jprot.2014.03.008
Gudipaty SA, D’Orso I (2016) Functional interplay between PPM1G and the transcription elongation machinery. RNA Disease 3(1):e1215
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E et al (2007) The 20 years of PROSITE. Nucleic Acids Res 36(Database):D245–D249. https://doi.org/10.1093/nar/gkm977
Jiang L, Chen Y, Luo L, Peck SC (2018) Central roles and regulatory mechanisms of dual-specificity MAPK phosphatases in developmental and stress signaling. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01697
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Lammers T, Lavi S (2007) Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit Rev Biochem Mol Biol 42:437–461. https://doi.org/10.1080/10409230701693342
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44(W1):W242–W245. https://doi.org/10.1093/nar/gkw290
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46(D1):D493–D496. https://doi.org/10.1093/nar/gkx922
Liu J, Stevens PD, Eshleman NE, Gao T (2013) Protein phosphatase PPM1G regulates protein translation and cell growth by dephosphorylating 4E binding protein 1 (4E-BP1). J Biol Chem 288(32):23225–23233. https://doi.org/10.1074/jbc.M113.492371
Mamoun CB, Sullivan DJ Jr, Banerjee R, Goldberg DE (1998) Identification and characterization of an unusual double serine/threonine protein phosphatase 2C in the malaria parasite Plasmodium falciparum. J Biol Chem 273:11241–11247. https://doi.org/10.1074/jbc.273.18.11241
Mamoun CB, Goldberg DE (2001) Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor 1β and inhibits its PKC-mediated nucleotide exchange activity in vitro: PP2C-mediated control of PfEF-1 β activity. Mol Microbiol 39(4):973–981. https://doi.org/10.1046/j.1365-2958.2001.02289.x
Mauzy MJ, Enomoto S, Lancto CA, Abrahamsen MS, Rutherford MS (2012) The Cryptosporidium parvum transcriptome during in vitro development. PLoS ONE 7(3):e31715. https://doi.org/10.1371/journal.pone.0031715
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203. https://doi.org/10.1093/nar/gkw1129
Mitula F, Tajdel M, Ciesla A, Kasprowicz-Maluski A, Kulik A, Babula-Skowronska D, Michalak M, Dobrowolska G, Sadowski J, Ludwikow A (2015) Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol 56:2351–2367. https://doi.org/10.1093/pcp/pcv146
Moorhead GB, Trinkle-Mulcahy L, Ulke-Lemee A (2007) Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol 8:234–244. https://doi.org/10.1038/nrm2126
Murray MV, Kobayashi R, Krainer AR (1999) The type 2C Ser/Thr phosphatase PP2Cγ is a pre-mRNA splicing factor. Genes Dev 13(1):87–97. https://doi.org/10.1101/gad.13.1.87
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Puiu D, Enomoto S, Buck GA, Abrahamsen MS, Kissinger JC (2004) CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res 32:D329–D331. https://doi.org/10.1093/nar/gkh050
Rajiv C, Davis TL (2018) Structural and functional insights into human nuclear cyclophilins. Biomolecules. https://doi.org/10.3390/biom8040161
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9:236–243. https://doi.org/10.1016/j.tplants.2004.03.007
Sela I, Ashkenazy H, Katoh K, Pupko T (2015) GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res 43(W1):W7–W14. https://doi.org/10.1093/nar/gkv318
Suh E-J, Kim T-Y, Kim SH (2006) PP2Cγ-mediated S-phase accumulation induced by the proteasome-dependent degradation of p21 WAF1/CIP1. FEBS Lett 580(26):6100–6104. https://doi.org/10.1016/j.febslet.2006.10.005
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6(7):e21800. https://doi.org/10.1371/journal.pone.0021800
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evolut 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Tewari R, Dorin D, Moon R, Doerig C, Billker O (2005) An atypical mitogen-activated protein kinase controls cytokinesis and flagellar motility during male gamete formation in a malaria parasite. Mol Microbiol 58(5):1253–1263. https://doi.org/10.1111/j.1365-2958.2005.04793.x
Travis SM, Welsh MJ (1997) PP2Cγ: a human protein phosphatase with a unique acidic domain 1. FEBS Lett 412(3):415–419. https://doi.org/10.1016/s0014-5793(97)00837-5
Wang B, Zhang P, Wei Q (2008) Recent progress on the structure of Ser/Thr protein phosphatases. Sci China Ser C Life Sci Chin Acad Sci 51:487–494. https://doi.org/10.1007/s11427-008-0068-y
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinform 9(1):40. https://doi.org/10.1186/1471-2105-9-40
Acknowledgements
This work is one of the requirements for Jenny Nancy Gómez Sandoval to obtain a PhD degree in Posgrado en Ciencias Biológicas (UNAM). She deeply appreciates the doctoral fellowship from CONACyT, Mexico (fellowship 203485, CVU 205974). The authors gratefully acknowledge technical assistance provided by Marco Gudiño Zayas from the Unidad de Investigación en Medicina Experimental, Facultad de Medicina, UNAM. We are thankful for technical support given by Mónica Mondragón from the Biochemistry Department and Electron Microscopy Facility-LANSE at CINVESTAV, Mexico.
Funding
This project was partially supported by Grants 152433 and 284018 from SEP-Consejo Nacional de Ciencia y Tecnología (SEP-CONACyT, Mexico) and Grant IN218619 from DGAPA-PAPIIT given to MMAG.
Author information
Authors and Affiliations
Contributions
MMA-G designed the study and revised the manuscript. PO reviewed the final manuscript. RM-F performed the microscopic analysis. ARE-M carried out Western blot assay. JNG-S performed the in-silico analysis and wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gómez-Sandoval, J.N., Okhuysen, P., Mondragón-Flores, R. et al. Cellular Identification and In Silico Characterization of Protein Phosphatase 2C (PP2C) of Cryptosporidium parvum. Acta Parasit. 65, 704–715 (2020). https://doi.org/10.2478/s11686-020-00209-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11686-020-00209-y