Abstract
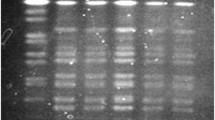
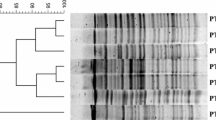
Cholera due to Vibrio cholerae has been spreading worldwide, although the reports focusing on Indonesian V. cholerae are few. In this study, in order to investigate how V. cholerae transmitted to human from environment. We extended an epidemiological report that had investigated the genotype of V. cholerae isolated from human pediatric samples and environmental samples. We examined 44 strains of V. cholerae isolated from pediatric diarrhea patients and the environment such as shrimps or oysters collected in three adjacent towns in Surabaya, Indonesia. Susceptibilities were examined for 11 antibiotics. Serotype O1 or O139 genes and pathogenic genes including cholera toxin were detected. Multi-locus sequence typing (MLST) and enterobacterial repetitive intergenic consensus (ERIC)-PCR were also performed to determine genetic diversity of those isolates. Serotype O1 was seen in 17 strains (38.6%) with all pathogenic genes among 44 isolates. Other isolates were non-O1/non-O139 V. cholerae. Regarding antibiotic susceptibilities, those isolates from environmental samples showed resistance to ampicillin (11.4%), streptomycin (9.1%) and nalidixic acid (2.3%) but those isolates from pediatric stools showed no resistance to those 3 kinds of antibiotics. MLST revealed sequence type (ST) 69 in 17 strains (38.6%), ST198 in 3 strains (6.8%) and non-types in 24 strains (54.5%). All the ST69 strains were classified to O1 type with more than 95% similarity by ERIC-PCR, including all 6 (13.6%) isolates from environmental samples with resistance to streptomycin. In conclusion, V. cholerae O1 ST69 strains has been clonally spreading in Surabaya, exhibiting pathogenic factors and antibiotic resistance to streptomycin, especially in the isolates from environment.

Similar content being viewed by others
References
Hartley DM, Morris JG Jr, Smith DL (2006) Hyperinfectivity: a critical element in the ability of V. cholerae to cause epidemics? PLOS Med 3:e7
Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, FreemanM Joyce K, Turnsek M, Garrett N, Humphrys M, Gomez G, Stroika S, Boncy J, Ochieng B, Oundo J, Klena J, Smith A, Keddy K, Smidt PG (2011) Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis 17:2122–2129
Rashed SM, Mannan SB, Johura FT, Islam MT, Sadique A, Watanabe H, Sack RB, Huq A, Colwell RR, Cravioto A, Alam M (2012) Genetic characteristics of drug-resistant Vibrio cholerae O1 causing endemic cholera in Dhaka, 2006–2011. J Med Microbiol 61:1736–1745
World Health Organization (2016) Health topics; Cholera. http://www.who.int/topics/cholera/en/. Accessed 9 Oct 2016
Faruque SM, Albert MJ, Mekalanos JJ (1998) Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev 62:1301–1314
Voss E, Manning PA, Attridge SR (1996) The toxin-coregulated pilus is a colonization factor and protective antigen of Vibrio cholerae El Tor. Microb Pathog 20:141–153
Rivera IN, Chun J, Huq A, Sack RB, Colwell RR (2001) Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl Environ Microbiol 67:2421–2429
Singh DV, Isac SR, Colwell RR (2002) Development of a hexaplex PCR assay for rapid detection of virulence and regulatory genes in Vibrio cholerae and Vibrio mimicus. J Clin Microbiol 40:4321–4324
Thelin KH, Taylor RK (1996) Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun 64:2853–2856
Chomvarin C, Johura FT, Mannan SB, Jumroenjit W, Kanoktippornchai B, Tangkanakul W, Tantisuwichwong N, Huttayananont S, Watanabe H, Hasan NA, Huq A, Cravioto A, Colwell RR, Alam M (2013) Drug response and genetic properties of Vibrio cholerae associated with endemic cholera in north-eastern Thailand, 2003–2011. J Clin Microbiol 62:599–609
Jun LJ, Kim JH, Jin JW, Jeong HD (2012) Characterization of a new beta-lactamase gene from isolates of Vibrio spp. in Korea. J Microbiol Biotech 22:555–562
Dalsgaard A, Forslund A, Sandvang D, Arntzen L, Keddy K (2001) Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J Antimicrob Chemother 48:827–838
Kutar BM, Rajpara N, Upadhyay H, Ramamurthy T, Bhardwaj AK (2013) Clinical isolates of Vibrio cholerae O1 El Tor Ogawa of 2009 from Kolkata, India: preponderance of SXT element and presence of Haitian ctxB variant. PLoS ONE 8:e56477
Kim HB, Wang M, Ahmed S, Park CH, LaRocque RC, Faruque AS, Salam MA, Khan WA, Qadri F, Calderwood SB, Jacoby GA, Hooper DC (2010) Transferable quinolone resistance in Vibrio cholerae. Antimicrob Agents Chemother 54:799–803
Baranwal S, Dey K, Ramamurthy T, Nair GB, Kundu M (2002) Role of active efflux in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob Agent Chemother 46:2676–2678
Abd El Ghany M, Chander J, Mutreja A, Rashid M, Hill-Cawthorne GA, Ali S, Naeem R, Thomson NR, Dougan G, Pain A (2014) The population structure of Vibrio cholerae from the Chandigarh Region of Northern India. PLoS Negl Trop Dis 8:e2981
Octavia S, Salim A, Kurniawan J, Lam C, Leung Q, Ahsan S, Reeves PR, Nair GB, Lan R (2013) Population structure and evolution of non-O1/non-O139 Vibrio cholerae by multilocus sequence typing. PLoS ONE 8:e65342
Waturangi DE, Joanito I, Yogi Y, Thomas S (2012) Use of REP- and ERIC-PCR to reveal genetic heterogeneity of Vibrio cholerae from edible ice in Jakarta, Indonesia. Gut Pathog 4:2
Esteves K, Mosser T, Aujoulat F, Hervio-Heath D, Monfort P, Jumas-Bilak E (2015) Highly diverse recombining populations of Vibrio cholerae and Vibrio parahaemolyticus in French Mediterranean coastal lagoons. Front Microbiol 6:708
Nishibori T, de Vries GC, Rahardjo D, Wasito EB, De I, Kinoshita S, Hayashi Y, Hotta H, Kawabata M, Shirakawa T, Iijima Y, Osawa R (2011) Phenotypic and genotypic characterization of Vibrio cholerae clinically isolated in Surabaya, Indonesia. Jpn J Infect Dis 64:7–12
Clinical and Laboratory Standards Institute (2014) Performance standards for antimicrobial susceptibility testing. In: 24th informational supplement. Clinical and Laboratory Standards Institute Document. M100-S24
Yasufuku T, Shigemura K, Shirakawa T, Matsumoto M, Nakano Y, Tanaka K, Arakawa S, Kinoshita S, Kawabata M, Fujisawa M (2011) Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli Strains clinically isolated from urinary tract infection patients. J Clin Microbiol 49:189–194
Nandi B, Nandy K, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC (2000) Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein ompW. J Clin Microbiol 38:4145–4151
Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, Nair GB, Shimada T, Takeda Y (1998) Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol 20:201–207
Yagi T, Kurokawa H, Shibata N, Shibayama K, Arakawa Y (2000) A preliminary survey of extended-spectrum beta-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol Lett 184:53–56
Ramazanzadeh R, Rouhi S, Shakib P, Shahbazi B, Bidarpour F, Karimi M (2015) Molecular characterization of Vibrio cholerae isolated from clinical samples in Kurdistan Province, Iran. Jundishapur J Microbiol 8:e18119
Ali M, Emch M, Donnay JP, Yunus M, Sack RB (2002) Identifying environmental risk factors for endemic cholera: a raster GIS approach. Health Place 8:201–210
Andersen JL, He GX, Kakarla P, Kc R, Kumar S, Lakra WS, Mukherjee MM, Ranaweera I, Shrestha U, Tran T, Varela MF (2015) Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int J Environ Res Public Health 12:1487–1547
Siriphap A, Leekitcharoenphon P, Kaas RS, Theethakaew C, Aarestrup FM, Sutheinkul O, Hendriksen RS (2017) Characterization and genetic variation of Vibrio cholerae isolated from clinical and environmental sources in Thailand. PLoS ONE 12:e0169324
Dalsgaard A, Mazur J, Dalsgaard I (2002) Misidentification of Vibrio cholerae O155 isolated from imported shrimp as O serogroup O139 due to cross-agglutination with commercial O139 antisera. J Food Protect 65:670–672
Anandan S, Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Thangamani S, Veeraraghavan B (2017) Prevailing clone (ST69) of Vibrio cholerae O139 in India over 10 years. Gut Pathog 9:60
Acknowledgements
This study was supported by following Grants: Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from the Ministry of Education, Culture, Sport, Science and Technology in Japan (Grant No. 18fm0108004h0004); Japan Agency for Medical Research and Development (AMED) (Grant No. JP18fk0108019); JSPS KAKENHI Grant No. 18K10045; Tahir Professorship from the University of Airlangga, Surabaya, Indonesia; The Institute of Tropical Disease, University of Airlangga, Surabaya, Indonesia; Indonesia-Japan Collaborative Research Center for Emerging and Re-emerging Infectious Diseases.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no financial conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Osawa, K., Shigemura, K., Kitagawa, K. et al. Difference of Phenotype and Genotype Between Human and Environmental: Isolated Vibrio cholerae in Surabaya, Indonesia. Indian J Microbiol 60, 230–238 (2020). https://doi.org/10.1007/s12088-020-00861-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-020-00861-y