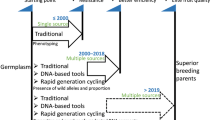
Abstract
Apple blue mold causes significant postharvest economic losses worldwide. A blue mold resistance locus, qM-Pe3.1, was previously identified on chromosome 3 of Malus sieversii PI 613981, a wild accession with inferior fruit quality. Introgression of the resistance allele into elite breeding germplasm is difficult and success of introgression and the effect of the PI 613981 genome on fruit quality cannot be phenotypically evaluated until fruiting, which occurs approximately 5 years from seed. In this study, introgression of the qM-Pe3.1 resistance allele was achieved by rapid cycle breeding, utilizing the transgenic line T1190 constitutively expressing the BpMADS4 early-flowering gene. This was supported by DNA-based diagnostic information that enabled marker-assisted selection for blue mold resistance using a locus-specific DNA test developed to detect the qM-Pe3.1 resistance allele in offspring (foreground selection). Of 75 second-generation ([‘Gala’ × PI 613981] × T1190) offspring carrying BpMADS4, 43 also carried the qM-Pe3.1 resistance allele. DNA tests for other trait loci were used to identify other desirable alleles related to fruit quality in progeny and 6874 genome-wide SNP markers (from an apple 20K Illumina® SNP array) were used to identify undesirable genomic segments of PI 613981 (background selection). Three individuals identified with favorable recombination close to qM-Pe3.1 and less than 25% of M. sieversii unimproved genome were selected as best suited for the elimination of unimproved DNA segments in subsequent generations. Our pipeline for introgression of qM-Pe3.1, facilitated by marker-assisted foreground and background selection, successfully advanced this promising germplasm in readiness for the next generation.







Similar content being viewed by others
References
Andersen B, Smedsgaard J, Frisvad JC (2004) Penicillium expansum: consistent production of patulin, chaetoglobosins, and other secondary metabolites in culture and their natural occurrence in fruit products. J Agric Food Chem 52:2421–2428
Aranzana M, Decroocq V, Dirlewanger E, Eduardo I, Gao Z, Gasic K, Iezzoni A, Jung S, Peace C, Prieto H, Tao R, Verde I, Abbott AG, Arús P (2019) Prunus genetics and applications after de novo genome sequencing: achievements and prospects. Hortic Res 6:58
Baumgartner IO, Kellerhals M, Costa F, Dondini L, Pagliarani G, Gregori R, Tartarini S, Leumann L, Laurens F, Patocchi A (2016) Development of SNP-based assays for disease resistance and fruit quality traits in apple (Malus × domestica Borkh.) and validation in breeding pilot studies. Tree Genet Genomes 12:1–21
Bianco L, Cestaro A, Sargent DJ, Banchi E, Derdak S, Guardo MD, Salvi S, Jansen J, Viola R, Gut I, Laurens F, Chagné D, Velasco R, van de Weg WE, Troggio M (2014) Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS One 9:e110377
Bianco L, Cestaro A, Linsmith G, Muranty H, Denancé C, Théron A, Poncet C, Micheletti D, Kerschbamer E, Pierro EAD, Larger S, Pindo M, van de Weg WE, Davassi A, Laurens F, Velasco R, Durel C-E, Troggio M (2016) Development and validation of the Axiom®Apple480K SNP genotyping array. Plant J 86:62–74
Bink MCAM, Jansen J, Madduri M, Voorrips RE, Durel C-E, Kouassi AB, Laurens F, Mathis F, Gessler C, Gobbin D, Rezzonico F, Patocchi A, Kellerhals M, Boudichevskaia A, Dunemann F, Peil A, Nowicka A, Lata B, Stankiewicz-Kosyl M, Jeziorek K, Pitera E, Soska A, Tomala K, Evans KM, Fernández-Fernández F, Guerra W, Korbin M, Keller S, Lewandowski M, Plocharski W, Rutkowski K, Zurawicz E, Costa F, Sansavini S, Tartarini S, Komjanc M, Mott D, Antofie A, Lateur M, Rondia A, Gianfranceschi L, van de Weg WE (2014) Bayesian QTL analyses using pedigreed families of an outcrossing species, with application to fruit firmness in apple. Theor Appl Genet 127:1073–1090
Bus V, Ranatunga C, Gardiner S, Bassett H, Rikkerink E, Geibel M, Fischer M, Fischer C (2000) Marker assisted selection for pest and disease resistance in the New Zealand apple breeding programme. Acta Hortic 538:541–547
Buti M, Poles L, Caset D, Magnago P, Fernandez Fernandez F, Colgan RJ, Velasco R, Sargent DJ (2015) Identification and validation of a QTL influencing bitter pit symptoms in apple (Malus × domestica). Mol Breed 35:29
Chagné D, Carlisle CM, Blond C, Volz RK, Whitworth CJ, Oraguzie NC, Crowhurst RN, Allan AC, Espley RV, Hellens RP (2007) Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics 8:212
Chagné D, Crowhurst RN, Troggio M, Davey MW, Gilmore B, Lawley C, Vanderzande S, Hellens RP, Kumar S, Cestaro A, Velasco R, Main D, Rees JD, Iezzoni A, Mockler T, Wilhelm L, van de Weg WE, Gardiner SE, Bassil N, Peace C (2012) Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS One 7:e31745
Cheng L, Jiang S, Zhang S, You H, Zhang J, Zhou Z, Xiao Y, Liu X, Du Y, Li J, Wang X, Xin Y, Zheng Y, Shang K (2016) Consumers’ behaviors and concerns on fresh vegetable purchase and safety in Beijing urban areas, China. Food Control 63:101–109
Cornille A, Gladieux P, Smulders MJ, Roldan-Ruiz I, Laurens F, Le Cam B, Nersesyan A, Clavel J, Olonova M, Feugey L, Gabrielyan I, Zhang XG, Tenaillon MI, Giraud T (2012) New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet 8:e1002703
Costa F, Stella S, van de Weg WE, Guerra W, Cecchinel M, Dallavia J, Koller B, Sansavini S (2005) Role of the genes Md-ACO1 and Md-ACS1 in ethylene production and shelf life of apple (Malus domestica Borkh). Euphytica 141:181–190
Daccord N, Celton J-M, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, Pierro EAD, Gouzy J, Rees DJG, Guérif P, Muranty H, Durel C-E, Laurens F, Lespinasse Y, Gaillard S, Aubourg S, Quesneville H, Weigel D, van de Weg WE, Troggio M, Bucher E (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49:1099–1106
de Capdeville G, Beer SV, Watkins CB, Wilson CL, Tedeschi LO, Aist JR (2003) Pre-and post-harvest harpin treatments of apples induce resistance to blue mold. Plant Dis 87:39–44
Di Pierro EA, Gianfranceschi L, Di Guardo M, Koehorst-van Putten HJ, Kruisselbrink JW, Longhi S, Troggio M, Bianco L, Muranty H, Pagliarani G, Tartarini S, Letschka T, Lozano Luis L, Garkava-Gustavsson L, Micheletti D, Bink MC, Voorrips RE, Aziz E, Velasco R, Laurens F, van de Weg WE (2016) A high-density, multi-parental SNP genetic map on apple validates a new mapping approach for outcrossing species. Hortic Res 3:16057
Dreesen RS, Vanholme BT, Luyten K et al (2010) Analysis of Malus S-RNase gene diversity based on a comparative study of old and modern apple cultivars and European wild apple. Mol Breed 26:693–709
Droby S, Wisniewski M, Macarisin D, Wilson C (2009) Twenty years of postharvest biocontrol research: is it time for a new paradigm? Postharvest Biol Technol 52:137–145
Edge-Garza D, Rowland T, Sandefur P, Konishi B, Brutcher L, Evans K, Watkins S, Oraguzie N, Clark M, Tillman J, Bedford D, Luby J, Peace C (2014a) Routine marker-assisted seedling selection focused on fruit quality improves breeding efficiency in three tree fruit programs. In: 7th International Rosaceae Genomics Conference. Seattle, WA, USA
Edge-Garza DA, Rowland TV, Haendiges S, Peace C (2014b) A high-throughput and cost-efficient DNA extraction protocol for the tree fruit crops of apple, sweet cherry, and peach relying on silica beads during tissue sampling. Mol Breed 34:2225–2228
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4:250–255
Evans K, Peace C (2017) Advances in marker-assisted breeding of apples. In: Evans K (ed) Achieving sustainable cultivation of apples. Burleigh Dodds Science Publishing, London, pp 189–216
Fischer C (1994) Shortening of the juvenile period in apple breeding. In: Schmidt H, Kellerhals M (eds) Progress in temperate fruit breeding. Springer, Dordrecht, pp 161–164
Flachowsky H, Peil A, Sopanen T, Elo A, Hanke V (2007) Overexpression of BpMADS4 from silver birch (Betula pendula Roth.) induces early-flowering in apple (Malus × domestica Borkh.). Plant Breed 126:137–145
Flachowsky H, Hanke M-V, Peil A, Strauss SH, Fladung M (2009) A review on transgenic approaches to accelerate breeding of woody plants. Plant Breed 128(3):217–226
Flachowsky H, Le Roux P, Peil A, Patocchi A, Richter K, Hanke M (2011) Application of a high-speed breeding technology to apple (Malus × domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytol 192:364–377
Forsline PL, Aldwinckle HS, Dickson EE, Luby JJ, Hokanson SC (2003) Collection, maintenance, characterization, and utilization of wild apples of Central Asia. Hortic Rev 29:1–61
Gu Z, Gu L, Eils R, Schlesner M, Brors B (2014) Circlize implements and enhances circular visualization in R. Bioinformatics 30:2811–2812
Guan Y, Peace C, Rudell D, Verma S, Evans K (2015) QTLs detected for individual sugars and soluble solids content in apple. Mol Breed 35:135
Janisiewicz WJ, Usall J, Bors B (1992) Nutritional enhancement of biocontrol of blue mold on apples. Phytopathology 82:1364–1370
Janisiewicz WJ, Saftner RA, Conway WS, Forsline PL (2008) Preliminary evaluation of apple germplasm from Kazakhstan for resistance to postharvest blue mold in fruit caused by Penicillium expansum. HortScience 43:420–426
Jung S, Lee T, Cheng C-H, Buble K, Zheng P, Yu J, Humann J, Ficklin SP, Gasic K, Scott K, Frank M, Ru S, Hough H, Evans K, Peace C, Olmstead M, DeVetter LW, McFerson J, Coe M, Wegrzyn JL, Staton ME, Abbott AG, Main D (2019) 15 years of GDR: new data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res 47:D1137–D1145
Jurick WM, Janisiewicz WJ, Saftner RA, Vico I, Gaskins VL, Park E, Forsline PL, Fazio G, Conway WS (2011) Identification of wild apple germplasm (Malus spp.) accessions with resistance to the postharvest decay pathogens Penicillium expansum and Colletotrichum acutatum. Plant Breed 130:481–486
Kassambara A (2017) ggpubr:“ggplot2” based publication ready plots. R Package Version 012
King GJ, Lynn J, Dover C, Evans K, Seymour G (2001) Resolution of quantitative trait loci for mechanical measures accounting for genetic variation in fruit texture of apple (Malus pumila Mill.). Theor Appl Genet 102:1227–1235
Konstantinou S, Karaoglanidis GS, Bardas GA, Minas IS, Doukas E, Markoglou AN (2011) Postharvest fruit rots of apple in Greece: pathogen incidence and relationships between fruit quality parameters, cultivar susceptibility, and patulin production. Plant Dis 95:666–672
Kotoda N, Iwanami H, Takahashi S, Abe K (2006) Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J Am Soc Hortic Sci 131:74–81
Kumar S, Garrick DJ, Bink MC, Whitworth C, Chagné D, Volz RK (2013) Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genomics 14:393
Le Roux P-M, Flachowsky H, Hanke M-V, Gessler C, Patocchi A (2012) Use of a transgenic early flowering approach in apple (Malus × domestica Borkh.) to introgress fire blight resistance from cultivar Evereste. Mol Breed 30:857–874
Liu J, Wisniewski M, Droby S, Tian S, Hershkovitz V, Tworkoski T (2011) Effect of heat shock treatment on stress tolerance and biocontrol efficacy of Metschnikowia fructicola. FEMS Microbiol Ecol 76:145–155
Longhi S, Hamblin MT, Trainotti L, Peace CP, Velasco R, Costa F (2013) A candidate gene based approach validates Md-PG1 as the main responsible for a QTL impacting fruit texture in apple (Malus × domestica Borkh). BMC Plant Biol 13:37
Luby JJ, Shaw DV (2001) Does marker-assisted selection make dollars and sense in a fruit breeding program? HortScience 36:872–879
Luo F, Evans K, Norelli JL, Zhang Z, Peace C (2020) Prospects for achieving durable disease resistance with elite fruit quality in apple breeding. Tree Genet Genomes 16:21
Mari M, Martini C, Spadoni A, Rouissi W, Bertolini P (2012) Biocontrol of apple postharvest decay by Aureobasidium pullulans. Postharvest Biol Technol 73:56–62
Morales H, Sanchis V, Usall J, Ramos AJ, Marín S (2008) Effect of biocontrol agents Candida sake and Pantoea agglomerans on Penicillium expansum growth and patulin accumulation in apples. Int J Food Microbiol 122:61–67
Mundt CC (2014) Durable resistance: a key to sustainable management of pathogens and pests. Infect Genet Evol 27:446–455
Norelli JL, Wisniewski M, Droby S (2013) Identification of a QTL for postharvest disease resistance to Penicillium expansum in Malus sieversii. Acta Hortic 1053:199–203
Norelli JL, Wisniewski M, Fazio G, Burchard E, Gutierrez B, Levin E, Droby S (2017) Genotyping-by-sequencing markers facilitate the identification of quantitative trait loci controlling resistance to Penicillium expansum in Malus sieversii. PLoS One 12:e0172949
Peace C, Piaskowski J, Vanderzande S (2018) Visualizing the genetics of elite genomes. In: 9th International Rosaceae Genomics Conference. Nanjing, China, 2018
Peace CP, Bianco L, Troggio M, van de Weg WE, Howard NP, Cornille A, Durel C-E, Myles S, Migicovsky Z, Schaffer RJ, Costes E, Fazio G, Yamane H, van Nocker S, Gottschalk C, Costa F, Chagné D, Zhang X, Patocchi A, Gardiner SE, Hardner C, Kumar S, Laurens F, Bucher E, Main D, Jung S, Vanderzande S (2019) Apple whole genome sequences: recent advances and new prospects. Hortic Res 6:59
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria 2016
Rosenberger DA (1997) Recent research and changing options for controlling postharvest decays of apples. In: Proceedings of harvesting, handling, and storage workshop, Northeast Regional Agricultural Engineering Service Publication NRAES-112. Cornell University, Ithaca
Rubio F, Alonso A, García-Martínez S, Ruiz JJ (2016) Introgression of virus-resistance genes into traditional Spanish tomato cultivars (Solanum lycopersicum L.): effects on yield and quality. Sci Hortic 198:183–190
Sandefur P, Oraguzie N, Peace C (2016) A DNA test for routine prediction in breeding of sweet cherry fruit color, Pav-Rf-SSR. Mol Breed 36:1–11
Schaller A, Vanderzande S, Peace C (2018) Fruit quality trait locus genotypes in apple from RosBREED’s SNP array data. In: Abstracts of the American Society for Horticultural Science annual meeting, July 30-August 3, Washington DC, USA
Schlathölter I, Jänsch M, Flachowsky H, Broggini GAL, Hanke M-V, Patocchi A (2018) Generation of advanced fire blight-resistant apple (Malus × domestica) selections of the fifth generation within 7 years of applying the early flowering approach. Planta 247:175–1488
Serra O, Donoso JM, Picañol R, Batlle I, Howad W, Eduardo I, Arús P (2016) Marker-assisted introgression (MAI) of almond genes into the peach background: a fast method to mine and integrate novel variation from exotic sources in long intergeneration species. Tree Genet Genomes 12:96
Sobel E, Papp JC, Lange K (2002) Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70:496–508
Spadoni A, Guidarelli M, Phillips J, Mari M, Wisniewski M (2015) Transcriptional profiling of apple fruit in response to heat treatment: involvement of a defense response during Penicillium expansum infection. Postharvest Biol Technol 101:37–48
Spotts RA, Cervantes LA, Mielke EA (1999) Variability in postharvest decay among apple cultivars. Plant Dis 83:1051–1054
Stegmeir T, Cai L, Basundari FRA, Sebolt AM, Iezzoni AF (2015) A DNA test for fruit flesh color in tetraploid sour cherry (Prunus cerasus L.). Mol Breed 35:149
Tahir I, Nybom H, Ahmadi-Afzadi M, Røen K, Sehic J, Røen D (2015) Susceptibility to blue mold caused by Penicillium expansum in apple cultivars adapted to a cool climate. Eur J Hortic Sci 80:117–127
Tanksley SD (1983) Molecular markers in plant breeding. Plant Mol Biol Rep 1:3–8
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115–e115
USDA, ARS, National Genetic Resources Laboratory (2019) Germplasm Resources Information Network (GRIN). https://npgsweb.ars-grin.gov/gringlobal/descriptors.aspx. Accessed 30 May 2019
Vanderzande S, Cai L, Howard NP, Da Silva LC, Antanaviciute L, Bink MCAM, Kruisselbrink JW, Bassil N, Gasic K, Iezzoni A, van de Weg WE, Peace C (2019) High-quality, genome-wide SNP genotypic data for pedigreed germplasm of the diploid outbreeding species apple, peach, and sweet cherry through a common workflow. PLoS One 14:e0210928
Vanderzande S, Piaskowski JL, Luo F, Edge-Garza DA, Klipfel J, Schaller A, Martin S, Peace C (2018) Crossing the finish line: how to develop diagnostic DNA tests as breeding tools after QTL discovery. J Hortic 5:1–6
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouze P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R (2010) The genome of the domesticated apple (Malus × domestica Borkh). Nat Genet 42:833–839
Verdu CF, Guyot S, Childebrand N, Bahut M, Celton J-M, Gaillard S, Lasserre-Zuber P, Troggio M, Guilet D, Laurens F (2014) QTL analysis and candidate gene mapping for the polyphenol content in cider apple. PLoS One 9:e107103
Verma S, Evans K, Guan Y, Luby JJ, Rosyara UR, Howard NP, Bassil N, Bink MCAM, van de Weg WE, Peace CP (2019) Two large-effect QTLs, Ma and Ma3, determine genetic potential for acidity in apple fruit: breeding insights from a multi-family study. Tree Genet Genomes 15:18
Volk GM, Chao CT, Norelli JL, Brown SK, Fazio G, Peace C, McFerson J, Zhong G-Y, Bretting P (2015) The vulnerability of US apple (Malus) genetic resources. Genet Resour Crop Evol 62:765–794
Volz RK, Rikkerink E, Austin P, Lawrence T, Bus VGM, Alonso JM (2009) “Fast-breeding” in apple: a strategy to accelerate introgression of new traits into elite germplasm. Acta Hortic 814:163–168
Weber RWS, Palm G (2010) Resistance of storage rot fungi Neofabraea perennans, N. alba, Glomerella acutata and Neonectria galligena against thiophanate-methyl in northern German apple production. J Plant Dis Prot 117:185–191
Wickham H (2010) ggplot2: elegant graphics for data analysis. J Stat Softw 35:65–88
Wisniewski M, Droby S, Norelli J, Liu J, Schena L (2016) Alternative management technologies for postharvest disease control: the journey from simplicity to complexity. Postharvest Biol Technol 122:3–10
Yamagishi N, Sasaki S, Yamagata K, Komori S, Nagase M, Wada M, Yamamoto T, Yoshikawa N (2011) Promotion of flowering and reduction of a generation time in apple seedlings by ectopical expression of the Arabidopsis thaliana FT gene using the Apple latent spherical virus vector. Plant Mol Biol 75:193–204
Yamagishi N, Kishigami R, Yoshikawa N (2014) Reduced generation time of apple seedlings to within a year by means of a plant virus vector: a new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnol J 12:60–68
You FM, Huo N, Gu YQ, Luo M, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9:253
Acknowledgments
Roger Lewis, Timothy Artlip, and Phil Welser from the USDA-ARS Appalachian Fruit Research Laboratory, Kearneysville, WV, and Terry Rowland and Stijn Vanderzande of the WSU Tree Fruit Genotyping Lab (Peace program) are thanked for their technical support. FL thanks the China Scholarship Council for providing funds for pursuing his PhD degree. This work was also supported by the USDA National Institute of Food and Agriculture-Specialty Crop Research Initiative project, “RosBREED: Combining disease resistance with horticultural quality in new rosaceous cultivars” (2014-51181-22378) and USDA NIFA Hatch project 1014919.
Data archiving statement
All SNP array data and results of fruit quality DNA testing are provided in Table S1; data regarding foreground selection for MBC1 offspring are included in Table S2; details about the five candidate SSR markers targeting the qM-Pe3.1 locus are provided in Table S3.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Chagné
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Luo, F., Norelli, J.L., Howard, N.P. et al. Introgressing blue mold resistance into elite apple germplasm by rapid cycle breeding and foreground and background DNA-informed selection. Tree Genetics & Genomes 16, 28 (2020). https://doi.org/10.1007/s11295-020-1419-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-020-1419-5