Abstract
Ashes (Fraxinus spp.) are important hardwood tree species in rural, suburban, and urban forests of the eastern USA. Unfortunately, emerald ash borer (EAB, Agrilus planipennis) an invasive insect pest that was accidentally imported from Asia in the late 1980s–early 1990s is destroying them at an alarming rate. All North American ashes are highly susceptible to EAB, although blue ash (F. quadrangulata) may have some inherent attributes that provide it some protection. In contrast Manchurian ash (F. mandshurica) is relatively resistant to EAB having coevolved with the insect pest in its native range in Asia. Given its level of resistance, Manchurian ash has been considered for use in interspecies breeding programs designed to transfer resistance to susceptible North American ash species. One prerequisite for successful interspecies breeding is consistency in chromosome ploidy level and number between the candidate species. In the current study, we cytologically determined that both Manchurian ash and blue ash are diploids (2n) and have the same number of chromosomes (2n = 2x = 46). We also characterized these species’ ribosomal gene families (45S and 5S rDNA) using fluorescence in situ hybridization (FISH). Both Manchurian and blue ash showed two 45S rDNA and one 5S rDNA sites, but blue ash appears to have an additional site of 45S rDNA. The 5S rDNA in both species is colocalized interstitially with one of the 45S rDNA sites. The copy number of these two ribosomal gene families in Manchurian ash were observed to be quite varied, which indicates the species are still undergoing evolutionary homogenization.
Similar content being viewed by others
Introduction
Ashes (Fraxinus spp.) are important forest and landscape trees worldwide. They have a diploid chromosome number of 2n = 2x = 46, but some ploidy variation has been reported (between species and within some species), including tetraploids (4n = 92), hexaploids (6n = 138), and octaploids (8n = 184) (Wright 1944; Santamour Jr 1962; Schaefer and Miksche 1977; Black and Beckman 1983; Whittemore et al. 2018). Two ash species, white (F. americana L.) and green (F. pennsylvanica Marsh.), are the most widely distributed and grown in the USA. White ash is most often found on moist upland sites, while green ash is found on bottomland sites such as flood plains of rivers and swampy areas, although some overlap of niche distribution can be found. Economically the North American ashes are important forest trees in the northeastern and mid-western USA where they grow from Maine to Florida and west to and through the Great Plains. Ash is considered to be a quality hardwood, and the wood is used for sports items such as baseball bats and musical instruments such as guitars, as well as furniture, cabinets, and boxes for packing and shipping.
The North American ashes are being killed by an invasive insect, the emerald ash borer (EAB, Agrilus planipennis Fair.), at an alarming rate since being first reported in 2002 in Michigan (EABIN 2010; Kovacs et al. 2010). Generally, the symptoms are not noticed until they become severe, typically noted by crown dieback. The damage from EAB could be devastating ecologically and economically, with an estimated cost to communities in billions of dollars (Kovacs et al. 2010; Sydnor et al. 2011). The EAB is believed to have been introduced accidentally to the USA in infested solid wood packing material from Asia during the late 1980s to mid-1990s (Cappaert et al. 2005; Siegert et al. 2008). The invasive insect has killed tens of millions of ash trees in the USA and threatens some eight billion ash trees in North America if measures are not taken to control the pest (Sydnor et al. 2007; Poland et al. 2015). As of October 2018, EAB has spread to 35 US states and the Canadian provinces of Ontario, Québec, New Brunswick, Nova Scotia, and Manitoba (EABIN 2019).
Closely related interfertile plant species are a potential source of valuable genetic variation for traits such as growth rate, wood quality, and resistance to pests and pathogens through interspecies hybridization. All native North American ash species are found to be susceptible to EAB, with blue ash (F. quadrangulata Michx.) showing a less susceptible response to EAB, possibly due to reduced host preference (Pureswaran and Poland 2009; Peterson et al. 2015). In contrast, Manchurian ash (F. mandshurica Rupr), a native to East Asia, is resistant to EAB as a result of it coevolving with the insect (Zhao et al. 2005), and as such is considered a potential source of EAB resistance for use in breeding with North American ashes.
Prior knowledge of species ploidy levels is a predictor of interspecies hybrid success for introgression of novel characters from a donor species in hybrid breeding programs. Cytological analysis to determine the ploidy level by counting the chromosome number from root to tip meristems is accurate and clearly demonstrates the species ploidy level (Mock et al. 2012; Sakhanokho and Islam-Faridi 2014). Additionally, the cytological analysis of chromosome number of a species can further determine whether the species is an aneuploid (variation of chromosome number from diploid) and reveal whether large structural components of the genome are similar or different between related species. Such information is useful in planning and implementing interspecies breeding programs.
Though the chromosome numbers of Manchurian ash and blue ash have been cataloged as 2n = 48 and 2n = 46, respectively (Index to Plant Chromosome Numbers. Missouri Botanical Garden, St. Louis. http://www.tropicos.org/Project/IPCN), no details describing the methods or results can be found. We, therefore, undertook this present research to confirm the chromosome number of these two species using a standard enzymatic digestion technique of root tips (Jewell and Islam-Faridi 1994). In addition, since cyto-molecular characterization of repetitive gene families has not been reported using repetitive gene families in Manchurian ash and blue ash, we did so by characterizing the ribosomal DNA gene family (18S–26S and 5S rDNAs) using fluorescence in situ hybridization (FISH). Ribosomal DNAs have been used as FISH probes for evolutionary and phylogenetic relationships between species and serve as landmark markers for chromosome(s) identification (e.g., karyotyping) and genome organization studies of various plant species including trees (e.g., Brassica – Maluszynska and Heslop-Harrison 1993; Pinus – Doudrick et al. 1995; Cai et al. 2006; Quercus soecies – Zoldos et al. 1999; Picea abies – Vischi et al. 2003; Coffea – Hamon et al. 2009; Fagaceae – Ribeiro et al. 2011).
Materials and methods
Plant materials and chromosome preparation
Manchurian ash (Lawyer Nursery Lot #E179) and blue ash (Sheffield, Seed Lot #40190, provenance Wisconsin, USA) seedlings were grown in potting soil in a greenhouse at Texas A&M University in College Station, TX, USA. Actively growing root tips, about 1.5 cm long, were harvested into a saturated aqueous solution of α-bromonaphthalene (0.8%), placed in the dark for 2.5–3.0 h accumulate metaphases, and then fixed in 4:1 (95% ethanol:glacial acetic acid) solution. The root tips were then enzymatically digested to prepare chromosome spreads without cover glasses as described by Jewell and Islam-Faridi (1994), except that the enzyme solution was modified as follows: 40% (v/v) Cellulase (C2730, Sigma, St. Louis, MO, USA), 20% (v/v) Pectinase (P2611, Sigma), 40% (v/v) 0.01 M citrate buffer (trisodium citrate dihydrate) (Mallinckrodt Baker, Phillipsburg, NJ, USA), citric acid monohydrate, pH 4.5 (Fisher Scientific, USA), 2% (w/v) Cellulase RS (SERVA Electrophoresis GmbH, Heidelberg, Germany), 3% (w/v) Cellulase R10 (Yakult Pharmaceutical, Tokyo, Japan), 1% (w/v) Macerozyme (Yakult Pharmaceutical, Tokyo, Japan), and 1.5% (w/v) Pectolyase Y23 (Kyowa Chemical Co., Osaka, Japan). The chromosome spreads were checked with a phase-contrast microscope (AxioImager A1, Carl Zeiss, Inc., Germany), and slides containing good chromosome spreads were selected and stored at −80 °C for use in the FISH.
Probe DNA and fluorescence in situ hybridization (FISH)
The FISH procedure was conducted to determine the chromosomal location of 5S and 18S-5.8S-26S rDNA genes in Manchurian ash and blue ash. Whole plasmid DNA with a 18S-5.8S-26S rDNA insert of maize (Zea mays) (Zimmer et al. 1988) or a 5S rDNA insert of sugar beet (Beta vulgaris) (Schmidt et al. 1994) including the spacer region were labeled with biotin-16-dUTP (Biotin-Nick Translation Mix, Roche, Indianapolis, IN) or digoxigenin-11-dUTP (Dig-Nick-Translation Mix, Roche) following manufacturers’ instructions.
Standard FISH technique was utilized as previously reported, essentially following the procedure described by Islam-Faridi et al. (2009) and Sakhanokho and Islam-Faridi (2013). Sites of biotin-labeled probe hybridization were detected using Cy3-conjugated streptavidin (Jackson Immuno Research Laboratories, USA). Sites of digoxigenin-labeled probe hybridization were visualized using fluorescein-conjugated sheep anti-digoxigenin (Roche, Germany). A small drop (10 μl) of VECTASHIELD containing DAPI (Vector Laboratories, USA), to counterstain the chromosomes, was added to the preparation, and the slides were covered with a glass cover slip (50 × 24 mm) to prevent photobleaching of the fluorochromes and overflowing of immersion oil when checking the FISH results under the required magnification.
Digital image capture and process
FISH images were viewed under a 63X plan apochromatic oil-immersion objective, and digital images were recorded using an epifluorescence microscope (Axio Imager M2, Carl Zeiss Inc., Germany) with suitable filter sets (Chroma Technology, Bellows Falls, VT, USA) and captured with a CoolCube 1 (MetaSystems Group Inc., Boston, MA, USA) high performance charge-coupled device (CCD) camera. Images were pre-processed with Ikaros and ISIS v5.1 (MetaSystems Inc.) and then further processed with Adobe Photoshop CS v8 (Adobe Systems Inc., Broadway, NY, USA).
Results and discussion
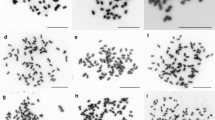
We successfully prepared the Manchurian ash and the blue ash somatic chromosome spreads from root meristems using an enzyme digestion technique (Jewell and Islam-Faridi 1994; Sakhanokho and Islam-Faridi 2013). DAPI-stained images showed that the chromosomes are mostly metacentric and submetacentric as revealed by their centromere positions, and both the species were diploids with 46 chromosomes (Fig. 1). The spreads are in unifocal position and are free of cell walls, nuclear membranes, cell debris–all critical prerequisites for successful FISH and accurate ploidy determination. Our results for blue ash (2n = 2x = 46) confirm the chromosome count reported in the Index to Plant Chromosome Numbers (Starodubtsev 1985). However, for the Manchurian ash we observed this accession to be diploid with 46 chromosomes and not 2n = 2x = 48 as reported in the Index. It is not uncommon for the nucleolus organizer regions (NORs) to become detached from their chromosomes during chromosome spreading resulting in a chromosome counts that are too high. Further, we have found that if the NOR is close to the centromere and the short arm of their chromosome is substantially larger than NOR’s associated satellite, it is more likely to detach. This has been observed in sorghum (Sorghum bicolor) and lentil (Lens culinaris) (Islam-Faridi – unpublished), and we suspect that it may be the case for the previously reported data for Manchurian ash.
FISH probed with 45S (green signals) and 5S rDNA (red signals) on Manchurian ash (left hand side panel, 1a to 1d) and blue ash (right hand side panel, 1e to 1h) chromosome spreads; 1a and 1e DAPI stained metaphase chromosome spreads; 1b and 1f 45S rDNA signals (green, arrow heads); 1c and 1g 5S rDNA signals (red, solid arrows); 1d and 1h superimposed image of 1a to 1c (Manchurian ash) and 1e to 1g (blue ash), respectively. The nucleolus organizing region (NOR) and the 45S rDNA site may sometimes detach (double arrow heads) from the original body of the chromosome as shown in ‶1f and 1h″. Scattered 45S and 5S rDNA FISH signals (green and red, respectively; in ‶1b to 1d″) can be seen in a partial interphase cell (marked with dotted light blue line). Heteromorphic FISH signals from 45S (green signals, arrow heads) and 5S (red signals, solid arrows) rDNAs on metaphase chromosome are clearly shown in the insert (enlarged image) in ‶1d″. Scale bar = 10 μm
We observed two sites of 18S-5.8S-26S rDNA (commonly known as 45S rDNA, and henceforth referred to as such) and one site of 5S rDNA in the Manchurian ash. One of the 45S rDNA sites is colocalized and intermingled with the 5S rDNA site, which is located interstitially in the short arm and at close proximity of the NOR (Fig. 2). One of the 45S rDNAs clearly showed a stronger FISH signal than the other (Figs. 1b, d, and 2). Furthermore, one of the 5S rDNA signals is much stronger than the others across the cell spreads and is always observed to be associated with a weaker 45S rDNA signal (Figs. 1d and 2). Strong FISH signals have been reported to be associated with higher rDNA copy number (Maluszynska and Heslop-Harrison 1993; Zoldos et al. 1999; Ribeiro et al. 2011), thus indicating the potential presence of 45S rDNA heteromorphism in these ash species. The other 45S rDNA signal pair that are not associated with the 5S rDNA also showed the heteromorphism, i.e., one 45S rDNA FISH signal is stronger than the other. The 5S and 45S rRNA genes in Manchurian ash were observed to be located at about the same position, that is, about halfway distal on the short arm (Fig. 2). We hypothesize this colocalization may be interspersed repeats of 5S and 45S rRNA genes as illustrated in the model shown in Fig. 2g. This colocalization and its organization can be proven by extended DNA fiber technique (Zoldos et al. 2018) and by molecular methods (Garcia et al. 2009). Recently Siljak-Yakovlev et al. (2014) reported similar results in F. ornus for numbers of 5S and 45S rDNA, and their colocalization, but they did not find any differential signal intensity for either 5S or 45S rDNA. Furthermore, the colocalization of 5S and 45S genes was quite different than our result in Manchurian ash. In F. ornus, the colocalization is confined in the satellite. We speculated that these two Fraxinus spp. could have originated from a common ancestor and the 5S gene location shifted from its original position in one of these species.
Two pairs of homologous chromosomes of a metaphase spread of Manchurian ash FISH with ribosomal rDNA probes; a) Green (45S rDNA) and red (5S rDNA) signals, b) 45S rDNA signals (green), cb1 and cb2) 5S rDNA signals (red) and d) DAPI-stained chromosomes (blue), putative centromeric position marked as arrows in bb1 and bb2, and NOR sites are marked as arrows in da1 to db2. Chromosomes in columns a1 and a2 are a homologous pair and same for columns b1 and b2. The overlapping region of 45S and 5S rDNA loci are marked (encircled) as white dotted lines (ab1 and ab2), and also shown in panel f and g. Diagrammatic representation of aa1-aa2 and ab1-ab2 of these homologues are shown in panels e and f, respectively. The hypothesized DNA dispersed repeat model is shown in g.
Heteromorphism of 5S rDNA is rare and is typically assumed to be constant (homomorphic) across species and even genera. Hetermorphism in both rDNA loci, that is, a strong 45S rDNA FISH signal (high copy number) is always associated with a weaker 5S rDNA (low copy number) and vice versa. To our knowledge, such observation is the first of its kind in any plant species. This heteromorphism in rDNA loci may suggest that Fraxinus spp. is still being evolutionarily homogenized. Alternatively, it could be a general feature of individual rDNA organization in Fraxinus similar to what has been shown in some other species (Zoldos et al. 1999; Ribeiro et al. 2011). Additional research in Fraxinus will be needed to determine which the case is or whether there is another explanation. In our FISH data, blue ash showed two, possibly three 45S rDNA and one 5S rDNA sites. Additional research is needed to confirm the number of 45S rDNA in blue ash. One of the 45S rDNA sites is clearly colocalized with the 5S rDNA site, which is located interstitially (Fig. 1h).
Plants’ hosts coevolve with sympatric pests and pathogens and consequentially can exhibit significant levels of resistance, but pests and pathogens introduced into non-native environments can cause enormous damage to the local, non-coevolved species (Ellison et al. 2005; Loo 2009). When a host tree species dominates a forest or urban environment, the destruction can be catastrophic costing millions of dollars to local and regional economies as well as causing forest ecosystem imbalance through loss of biodiversity (Dalgleish et al. 2016). For example, chestnut blight (Cryphonectria parasitica (Murrill.) Barr.) on American chestnut (Castanea dentata (Marsh.) Borkh.) (Hepting 1974; Kuhlman 1978), Dutch elm disease (Ophiostoma ulmi (Buisman) Melin & Nannf.) on American elm (Ulmus americana L.) (Hubbes 1999), mountain pine beetle (Dendroctonus ponderosae Hopkins) on various western conifers (Negrón and Fettig 2014), and more recently the North American ashes and European common ash (F. excelsior) are being destroyed by EAB and an ash dieback pathogen (Hymenoscyphus fraxineus, previously known as Chalara fraxinea), respectively. The ash dieback disease was first documented in early 1990s in the Baltic countries and eastern Poland (Przybyl 2002) and has now spread throughout Europe including the British Isles (MacLean et al. 2013; McKinney et al. 2014). As we know from the history of Dutch elm disease and chestnut blight, it is essentially impossible to stop an introduced, invasive pathogen from spreading across the species range. Ash dieback may destroy most of the ashes in Europe if no resistance is found in their local ashes. Furthermore, EAB has recently been documented in eastern European countries and is spreading at an alarming rate since the pest was first noted in Moscow, Russia (Kinver 2013; Orlova-Bienkowskaja 2014; Valenta et al. 2016). This could be another negative impact to the European common ash and presents a significant threat to the ashes in North America.
Prior information of chromosome number and ploidy level of a donor (resistant) species is not essential for interspecies hybrid breeding; however, cytological information is helpful as a predictor of crossability and a diagnostic for understanding species’ cross combination successes and failures. Based on our cytological findings, we concluded that Manchurian ash and blue ash are diploids with the same chromosome number and are cytogenetically similar to the common North American ashes. However, phylogenetic (Whitehill et al. 2011; Hinsinger et al. 2013) and crossing (Koch, unpublished data) studies indicate that only Manchurian ash and black ash (F. nigra) are close enough to hybridize. Interspecific hybrids often unveil structural chromosomal differences in wide crossing programs with the objective of transferring novel trait(s) from one species into another (Nelson et al. 2014). Cytological analysis of meiocytes of the interspecific F1 hybrids can clearly reveal structural differences, such as inversions and/or translocations, present in either species, and when present these structural differences can block or reduce fertility in hybrids (Burnham 1956, 1984; Jáuregui et al. 2001; Farré et al. 2012). A cytological analysis of meiocytes of the ultimate breeding product (or newly developed line) would clearly demonstrate the background chromosome behavior, and whether the chromosomes are completely homologous (Islam-Faridi 1988; Qi et al. 2007; Rabiza-Swider et al. 2010).
The Northern Research Station (NRS), US Forest Service, has archived Manchurian ash from China for potential use in interspecies breeding to transfer EAB resistance into North American ashes. An initial effort in crossing Manchurian ash with native North American ashes produced a number of small families including initial F1 families and backcross families that are under evaluation for EAB resistance (Koch, unpublished data). Since Manchurian ash has been identified as a host and likely coevolved species of the ash dieback fungus (Zhao et al. 2012), these materials may be useful for dieback resistance as well as EAB resistance.
Given the common problems with invasive pests and pathogens of ash in Europe and North America, collaborations between researchers working toward a common goal to save the European common ash and the North American ashes should be productive. Such collaborations would help to speed up the discovery, development, and deployment of resistance ash trees on both continents. Cooperative studies aimed at resolving the cytological subtleties between the North American, European, and Asian ashes would provide a good basis for collaboration and provide important information for the genetics and genomics programs aimed at developing pest and pathogen resistance.
References
Black CL, Beckman RL (1983) The variability of nuclear DNA and its implications for polyploidy in white ash (Fraxinus americana L.: Oleaceae). Am J Bot 70:1420–1423. https://doi.org/10.2307/2443433
Burnham CR (1956) Chromosomal interchanges in plants. Bot Rev 22:419–552. https://doi.org/10.1007/BF02872484
Burnham CR (1984) Discussion in cytogenetics. Burgess Publishing Company, St. Paul
Cai Q, Zhang D, LiuZ-L X-R, Wang X-R (2006) Chromosomal localization of 5S and 18S rDNA in five species of subgenus Strobus and their implications for genome evolution of Pinus. Ann Bot 97:715–722. https://doi.org/10.1093/aob/mc1030
Cappaert D, McCullough DG, Poland T, Siegert N (2005) Emerald ash borer in North America: a research and regulatory challenge. Am Entomol 51:152–165. https://doi.org/10.1093/ae/51.3.152
Dalgleish HJ, Nelson CD, Scrivani JA, Jacobs DF (2016) Consequences of shifts in abundance and distribution of American chestnut for restoration of a foundation forest tree. Forests 7(4):1–9. https://doi.org/10.3390/f7010004
Doudrick RL, Heslop-Harrison JS, Nelson CD, Schmidt T, Nance WL, Schwarzacher T (1995) Karyotype of slash pine (Pinus elliottii var. elliottii) using patterns of fluorescence in situ hybridization and fluorochrome banding. J Hered 86:289–296. https://doi.org/10.1093/oxfordjournals.jhered.a111583
EABIN (Emerald Ash Borer Information Network) (2010) USDA Forest Service and Michigan State University, Lansing. https://www.emeraldashborer.info/timeline.php
EABIN (Emerald Ash Borer Information Network) (2019) USDA Forest Service and Michigan State University, Lansing. https://www.emeraldashborer.info/
Ellison AM, Bank MS, Clinton BD, Clinton BD, ColburnEA EK, Ford CR, Foster DR, Kloeppel BD, Knoepp JD, Lovett GM, Mohan J, Orwig DA, Rodenhouse NL, SobczakWV SKA, Stone JK, Swan CM, Jill Thompson J, Betsy Von Holle BV, Webster JR (2005) Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front Ecol Environ 3:479–486. https://doi.org/10.1890/1540-9295(2005)003[0479:LOFSCF]2.0.CO;2
Farré A, Cuadrado A, Lacasa-Benito I, Cistué L, Schubert I, Comadran J, Jansen J, Romagosa I (2012) Genetic characterization of a reciprocal translocation present in a widely grown barley variety. Mol Breed 30(2):1109–1119. https://doi.org/10.1007/s11032-011-9698-z
Garcia S, Lim KY, Chester M, Garnatje T, Pellicer J, Vallès J, Leitch AR, Kovařík A (2009) Linkage of 35S and 5S rRNA genes in Artemisia (family Asteraceae): first evidence from angiosperms. Chromosoma 118:85–97. https://doi.org/10.1007/s00412-008-0179-z
Hamon P, Siljak-Yakovlev S, Srisuwan S, Robin O, Poncet V, Hamon S, de Kochko A (2009) Physical mapping of rDNA and heterochromatin in chromosomes of 16 Coffea species: a revised view of species differentiation. Chromosom Res 17:291–304. https://doi.org/10.1007/s10577-009-9033-2
Hepting GH (1974) Death of the American chestnut. J For Hist 18(3):61–67
Hinsinger DD, Basak J, Gaudeul M, Cruaud C, Bertolino P, Frascaria-Lacoste N, Bousquet J (2013) The phylogeny and biogeographic history of ashes (Fraxinus, Oleaceae) highlight the roles of migration and Vicariance in the diversification of temperate trees. PLoS One 8(11):e80431. https://doi.org/10.1371/journal.pone.0080431
Hubbes M (1999) The American elm and Dutch elm disease. For Chron 75(2):265–273
Islam-Faridi MN (1988) Genetical studies of grain protein and developmental characters in wheat. A dissertation submitted for the degree of doctor of philosophy to the University of Cambridge, Cambridge
Islam-Faridi MN, Nelson CD, DiFazio S, Gunter L, Tuskan GA (2009) Cytogenetic analysis of Populus trichocarpa – ribosomal DNA, telomere sequence, and marker-selected BACs. Cytogenet Genome Res 125(1):74–80. https://doi.org/10.1159/000218749
Jáuregui B, Vicente MC, Messeguer R, Felipe A, Bonnet A, Salesses G, Arús P (2001) A reciprocal translocation between ‘Garfi’ almond and ‘Nemared’ peach. Theor Appl Genet 102:1169–1176. https://doi.org/10.1007/s001220000511
Jewell DC, Islam-Faridi MN (1994) Details of a technique for somatic chromosome preparation and C-banding of maize. In: Freeling M, Walbot V (eds) The maize hand book, Springer-Verlag, pp 484–493
Kinver M (2013) Emerald ash borer beetle on the march across Europe. Environment reporter, BBC Science and Environment 23 October 2013, https://www.bbc.com/news/science-environment-24612322
Kovacs K, Haight R, McCullough DG, Mercader R, Siegert N, Liebhold A (2010) Cost of potential emeralds ash borer damage in U.S. communities 2009-2019. Ecol Econ 69:569–578. https://doi.org/10.1016/j.ecolecon.2009.09.004
Kuhlman EG (1978).The devastation of American chestnut by blight. In "Proceedings of the American chestnut symposium", Eds. W.L. MacDonald and F.C. Cech, West Virginia University Morgantown, West Virginia January 4-5, 1978, pp 1–3
Loo JA (2009) Ecological impacts of non–indigenous invasive fungi as forest pathogens. Biol Invasions 11:81–96. https://doi.org/10.1007/s10530-008-9321-3
MacLean D, Yoshida K, Edwards A, Crossman L, Clavijo B, Clark M, Swarbreck D, Bashton M, Chapman P, Gijzen M, Caccamo M, Downie A, Kamoun S, Saunders DGO (2013) Crowdsourcing genomic analyses of ash and ash dieback – power to the people. GigaScience 2:2. https://doi.org/10.1186/2047-217X-2-2
Maluszynska J, Heslop-Harrison JS (1993) Physical mapping of rDNA loci in Brassica species. Genome 36:774–781. https://doi.org/10.1139/g93-102
McKinney LV, Nielsen LR, Collinge DB, Thomsen IM, Hansen JK, Kjær ED (2014) The ash dieback crisis: genetic variation in resistance can prove a long-term solution. Plant Pathol 63:485–499. https://doi.org/10.1111/ppa.12196
Mock KE, Callahan CM, Islam-Faridi MN, Rai HS, Sanderson SC, Rowe CA, Ryel RJ, Shaw JD, Madritch MD, Gardner RS, Wolf PG (2012) Widespread triploidy in western north American aspen (Populus tremuloides). PLoS One 7(10):e4840. https://doi.org/10.1371/journal.pone.0048406
Negrón JF, Fettig CJ (2014) Mountain pine beetle, a major disturbance agent in US western coniferous forests: a synthesis of the state of knowledge. For Sci 60(3):409–413. https://doi.org/10.5849/forsci.13-169
Nelson CD, Powell WA, Merkle SA, Carlson JE, Hebard FV, Islam-Faridi MN, Staton ME, Georgi LL (2014) Chestnut. In: Ramawat K (ed) Tree biotechnology, chapter 1. CRC Press, Boca Raton, pp 3–35
Orlova-Bienkowskaja MJ (2014) Ashes in Europe are in danger: the invasive range of Agrilus planipennis in European Russia is expanding. Biol Invasions 16:1345–1349. https://doi.org/10.1007/s10530-013-0579-8
Peterson DL, Duan JJ, Yaninek JS, Ginzel MD, Sadof CS (2015) Growth of larval Agrilus planipennisi (Hymenoptera: Eulophidae) in blue ash (Fraxinus quadrangulata) and green ash (F. pennsylvanica). Environ Entomol 44(6):1512–1521. https://doi.org/10.1093/ee/nvv122
Poland TM, Chen Y, Koch J, Pureswaran D (2015) Review of emerald ash borer (Coleoptera: Buprestidae), life history, mating behaviours, host plant selection, and host resistance. Can Entomol 147:252–262. https://doi.org/10.4039/tce.2015.4
Przybyl K (2002) Fungi associated with necrotic apical parts of Fraxinus excelsior shoots. For Pathol 32:387–394. https://doi.org/10.1046/j.1439-0329.2002.00301.x
Pureswaran D, Poland TM (2009) Host selection and feeding preference of Agrilus planipennis (Coleoptera: Buprestidae) on ash (Fraxinus spp.). Environ Entomol 38(3):757–765. https://doi.org/10.1603/022.038.0328
Qi L, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosom Res 15:3–19. https://doi.org/10.1007/s10577-006-1108-8
Rabiza-Swider J, Brzezinski W, Lukaszewaski AJ (2010) Breeding behavior of chromosomes 1R cytogenetically engineered for breadmaking quality in hexaploid triticale. Crop Sci 50:808–814. https://doi.org/10.2135/cropsci2009.06.0349
Ribeiro T, Loureiro J, Santos C, Morais-Cecilio L (2011) Evolution of rDNA FISH patterns in the Fagaceae. Tree Genet Genomes 7:1113–1122. https://doi.org/10.1007/s11295-011-0399-x
Sakhanokho HF, Islam-Faridi MN (2013) Nuclear DNA content, base composition, and cytogenetic characterization of Christia obcordata. J Am Soc Hortic Sci 138(3):205–209. https://doi.org/10.21273/JASHS.138.3.205
Sakhanokho HF, Islam-Faridi MN (2014) Spontaneous autotetraploidy and its impact on morphological traits and pollen viability in Solanum aethiopicum. HortScience 49(8):997–1002. https://doi.org/10.21273/HORTSCI.49.8.997
Santamour FS Jr (1962) The relation between polyploidy and morphology in white and Baltimore ashes. Bull Torrey Bot Club 89:228–232. https://doi.org/10.2307/2483198
Schaefer VG, Miksche JP (1977) Microspectrophotometric determination of DNA per cell and polyploidy in Fraxinus americana L. Silve Genetica 26(5–6):184–192
Schmidt T, Schwarzacher T, Heslop-Harrison JS (1994) Physical mapping of rRNA genes by fluorescent in situ hybridization and structural analysis of 5S rRNA genes and intergenic spacer sequences in sugar-beet (Beta vulgaris L.). Theor Appl Genet 88:629–636. https://doi.org/10.1007/BF01253964
Siegert N, McCullough DG, Liebhold A, Telewski F (2008) Reconstruction of the establishment of emerald ash borer through dendrochronological analysis. In K. McManus and K. Gottschalk (eds.), proceedings, 19th U.S. Department of Agriculture interagency research forum on invasive species 2008; 8-11 January 2008, U.S. Department of Agriculture, Forest Service, northern Research Station: 70, Annapolis, MD
Siljak-Yakovlev S, Temunovic M, Robin O, Raquin C, Feascaria-Lacoste N (2014) Molecular-cytogenetic studies of ribosomal RNA genes and heterochromatin in three European Fraxinus species. Tree Genet Genomes 10:231–239. https://doi.org/10.1007/s11295-013-0654-4
Starodubtsev VN (1985) Chromosome numbers in the representatives of some families from the Soviet Far East. Bot Zhurn SSSR 70(2):275–277
Sydnor TD, Bumgardner M, Todd A (2007) The potential economic impacts of emerald ash borer (Agrilus planipennis) on Ohio, U.U., communities. Arboricult Urban For 33(1):48–54
Sydnor TD, Bumgardner M, Subburayalu S (2011) Community ash densities and economic impact potential of emerald ash borer (Agrilus planipennis) in four Midwestern states. Arboricult Urban For 33(1):84–89
Valenta V, Moser D, Kapeller S, Essl F (2016) A new forest pest in Europe: a review of emerald ash borer (Agrilus planipennis) invasion. J Appl Sil Entomol 141:507–526. https://doi.org/10.1111/jen.12369
Vischi M, Jurman I, Bianchi G, Morgante M (2003) Karyotype of Norway spruce by multicolor FISH. Theor Appl Genet 107:591–597. https://doi.org/10.1111/jen.12369
Whitehill JGA, Popova-Butler A, Green-Church KB, Koch JL, Herms DA, Bonello P (2011) Interspecific proteomic comparisons reveal ash phloem genes potentially involved in constitutive resistance to the emerald ash borer. PLoS One 6(9):e24863. https://doi.org/10.1371/journal.pone.0024863
Whittemore AT, Cambell JJN, Zheng-Lian X, Carlson CH, Atha D, Olsen RT (2018) Ploidy variation in Fraxinus L. (Oleaceae) of eastern North America: genome size diversity and taxonomy in a suddenly endangered genus. Int J Plant Sci 179(5):377–389. https://doi.org/10.1086/696688
Wright JW (1944) Genotypic variation in white ash. J For:489–495. https://doi.org/10.1093/jof/42.7.489
Zhao TH, Gao RT, Liu HP, Bauer LS, Sun LQ (2005) Host range of emerald ash borer, Agrilus planipennis Fairmaire, its damage and the countermeasures. Acta Entomol Sin 48(4):594–599
Zhao Y-J, Hosoya T, Baral H-O, Hosaka K, Kakishima M (2012) Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon 122:25–41. https://doi.org/10.5248/122.25
Zimmer EA, Jupe ER, Walbot V (1988) Ribosomal gene structure, variation and inheritance in maize and its ancestors. Genetics 120:1125–1136
Zoldos V, Papes D, Cerbah M, Panaud O, Besendorfer V, Siljak-Yakovlev S (1999) Molecular-cytogenetic studies of ribosomal genes and heterochromatin reveal conserved genome organization among 11 Quercus species. Theor Appl Genet 99:969–977. https://doi.org/10.1007/s001220051404
Zoldos V, Biruš I, Muratovic E, Šatović Z, Vojta A, Robin O, Pustahija F, Bogunić F, Vičić-Bočkor V, Siljak-Yakovlev S (2018) Epigenetic differentiation of natural populations of Lilium bosniacum associated with contrasting habitat conditions. Genome Biol Evol 10(1):291–303. https://doi.org/10.1093/gbe/evy010
Acknowledgments
We would like to thank Drs. A. Abbott (University of Kentucky), B. Berson (USDA-ARS), H. Sakhanokho (USDA-ARS), F. Raley (Texas A&M Forest Service), and G. Hodnett (Texas A&M) for their critical reading and valuable comments on the manuscript. We also thank Dr. Raley for the use of greenhouse facilities.
Data archiving statement
The new findings on chromosomes numbers will be incorporated in the CCDB (Chromosome Counts Database, http://ccdb.tau.ac.il) and the number of rDNA gene loci in the rDNA database (http://plantrdnadatabase.com).
Funding
Funding for this research was provided by the Southern Research Station and Northern Research Station (USDA Forest Service).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Z. Kaya
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Islam-Faridi, N., Mason, M.E., Koch, J.L. et al. Cytogenetics of Fraxinus mandshurica and F. quadrangulata: ploidy determination and rDNA analysis. Tree Genetics & Genomes 16, 26 (2020). https://doi.org/10.1007/s11295-020-1418-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-020-1418-6