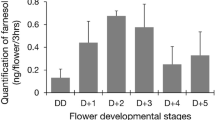
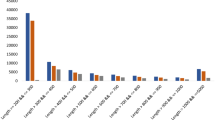
Abstract
The Chinese narcissus is a well-known monocotyledon plant with a beautiful color, and fresh with a sweet floral scent. Lack of transcriptomic and genomic information hinders understanding of the molecular mechanisms underlying the biosynthesis of narcissus floral scent volatiles. Here we predicted the functions of identified significantly differentially expressed genes (DEGs), according to public protein annotation databases. Using RNA-sequencing (RNA-Seq) on the Illumina HiSeq system and de novo transcriptome assembly, we investigated gene expression in narcissus corona and petal tissues at the early flowering (day 1) and full-bloom (day 7) stages. Significant differences in the expression profiles of 14 fragrance-related genes were further analyzed by qRT-PCR. A total of 62,826,860,514 bases were generated by RNA-seq; clean reads were 210,658,254 bp, and the guanine-cytosine content was 47.7%–48.88%. Transcripts (n = 167,374; 67.27%) and unigenes (n = 81,442; 32.73%) had mean lengths of 1069.70 bp and 813.27 bp, respectively. The total length and N50 length values of transcripts were 179,040,048 bp and 1654 bp, while those of unigenes were 66,234,291 bp and 1406 bp. Assembled genes were annotated by comparison with the non-redundant, Protein family, Clusters of Orthologous Groups of proteins, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes, and Gene Ontology, public protein databases. Additionally, 46 and 71 significantly differentially expressed genes encoded enzymes and transcription factors, respectively, associated with floral volatiles biosynthesis pathways, were analyzed in-depth. Our findings represent a fundamental step toward better understanding of the mechanisms of narcissus floral volatile biosynthesis.











Similar content being viewed by others
Abbreviations
- HS-SPME:
-
solid-phase microextraction
- GC-MS:
-
gas chromatography/mass spectrometry
- NES/LINS:
-
nerolidol/linalool synthase
- TPS:
-
terpene synthase
- AADC:
-
pyridoxal-50 -phosphate-dependent L-aromatic amino acid decarboxylase
- PAR:
-
phenylacetaldehyde reductase
- DXR:
-
1-deoxy-d-xylulose 5-phosphate reductoisomerase
- PAL:
-
phenylalanine ammonia-lyase
- BEAT:
-
benzyl alcohol acetyltransferase
- BAMT:
-
benzoic acid carboxyl methyl transferase
- SAMT:
-
salicylic acid carboxyl methyl- transferase
- HPL:
-
Hydroperoxide lyase
- 2AP:
-
2-acetyl-1-pyrroline
- BADH2 (AMADH2):
-
betaine aldehyde dehydrogenase 2
- NR:
-
non-redundant protein sequences
- Pfam:
-
Protein family
- KOG/COG/eggnog:
-
Clusters of Orthologous Groups of proteins
- Swiss-Prot:
-
a manually annotated and reviewed protein sequence database
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GO:
-
Gene Ontology
- DEGs:
-
differential expressed genes
- IPP:
-
isopentenyl diphosphate
- DMAPP (DMADP):
-
dimethylallyl diphosphate
- G3P:
-
glyceraldehyde 3-phosphate
- MEP:
-
2-C-methyl-D-erythritol 4-phosphate
- DXS:
-
1-D-desoxyxylulose 5-phosphate synthase
- DXP:
-
1-deoxy-d-xylulose 5-phosphate
- TPP:
-
thiamine pyrophosphate
- CMS:
-
2-C-methyl-D-erythritol 4- phosphate cytidylyltransferase
- CTP:
-
cytosine 5′-triphosphate
- ICME:
-
isoprenylcysteine methylesterase
- GGDP:
-
geranylgeranyl diphosphate
- Dedol-PP:
-
dehydrodolichyl diphosphate
- DDS:
-
dedol-PP synthase
- ABA:
-
abscisic acid
- GA:
-
gibberellin
- CDP-ME2P:
-
4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate
- ME-CPP:
-
2-C-methyl-D-erythritol 2,4-cyclodiphosphate
- MEcDP:
-
2- C-methyl-D-erythritol 2,4-cyclodiphosphate
- HMBPP:
-
1-hydroxy-2-methyl-butenyl 4-disphosphate
- FDP:
-
(E, E)-Famesyl-PP
- GDP:
-
Geranyl-PP
- DoI-PP:
-
Dolichol-PP
- MAS:
-
momilactone A synthase
- benzoyl-CoA:
-
O-benzoyltransferase
- DBAT:
-
10-deacetylbaccatin III 10-O-acetyltransferase
- Phe:
-
phenylalanine
- Tyr:
-
tyrosine
- TAL:
-
tyrosine ammonia lyase
- 4CL:
-
4-coumarate--CoA ligase
- ALDH:
-
aldehyde dehydrogenase
- CCR:
-
cinnamoyl CoA reductase
- CCoAOMT:
-
caffeoyl-CoA-O-methyltransferase
- CAD:
-
cinnamyl alcohol dehydrogenase
- ASAT:
-
aspartate aminotransferase
- HCT:
-
shikimate O-hydroxycinnamoyl transferase;
- benzoyl-CoA:
-
benzyl alcohol O-benzoyltransferase
- bZIP:
-
basic leucine zipper
- ERF:
-
ethylene-responsive factor
- bHLH:
-
basic helix-loop-helix
- SRF:
-
serum response factor
- TCP:
-
TEOSINTE-BRANCHED1/CYCLOIDEA/PCF
- EOBI:
-
EMISSION OF BENZENOIDS I
- MAPK:
-
mitogen-activated protein kinase
- JA:
-
jasmonate acid
- BIS1:
-
bHLH iridoid synthesis 1
- qRT-PCR:
-
real-time quantitative PCR
- FPKM:
-
fragments per kilobase of transcript per million mapped reads
- FDR:
-
false discovery rate
- TFs:
-
transcription factors
References
Anwar M, Wang G, Wu J, Waheed S, Allan A, Zeng L (2018) Ectopic overexpression of a novel R2R3-MYB, NtMYB2 from Chinese narcissus represses anthocyanin biosynthesis in tobacco. Molecules 23
Bai Z, Li W, Jia Y, Yue Z, Jiao J, Huang W, Xia P, Liang Z (2018) The ethylene response factor SmERF6 co-regulates the transcription of SmCPS1 and SmKSL1 and is involved in tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Planta 248:243–255
Banerjee A, Wu Y, Banerjee R, Li Y, Yan H, Sharkey TD (2013) Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J Biol Chem 288:16926–16936
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284
Bera P, Mukherjee C, Mitra A (2017) Enzymatic production and emission of floral scent volatiles in Jasminum sambac. Plant Sci 256:25–38
Bergström G, Dobson HEM, Groth I (1995) Spatial fragrance patterns within the flowers ofRanunculus acris (Ranunculaceae). Plant Syst Evol 195:221–242
Bohlmann J, Martin D, Oldham NJ, Gershenzon J (2000) Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-beta-ocimene synthase. Arch Biochem Biophys 375:261–269
Bouider N, Fhayli W, Ghandour Z, Boyer M, Harrouche K, Florence X, Pirotte B, Lebrun P, Faury G, Khelili S (2015) Design and synthesis of new potassium channel activators derived from the ring opening of diazoxide: study of their vasodilatory effect, stimulation of elastin synthesis and inhibitory effect on insulin release. Bioorg Med Chem 23:1735–1746
Bradbury LM, Fitzgerald TL, Henry RJ, Jin Q, Waters DL (2005) The gene for fragrance in rice. Plant Biotechnol J 3:363–370
Bresso EG, Chorostecki U, Rodriguez RE, Palatnik JF, Schommer C (2018) Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol 176:1694–1708
Celedon JM, Chiang A, Yuen MM, Diaz-Chavez ML, Madilao LL, Finnegan PM, Barbour EL, Bohlmann J (2016) Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J 86:289–299
Chen F, Tholl D, D'Auria JC, Farooq A, Pichersky E, Gershenzon J (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15:481–494
Chen HC, Chi HS, Lin LY (2013) Headspace solid-phase microextraction analysis of volatile components in Narcissus tazetta var. chinensis Roem. Molecules 18:13723–13734
Cunillera N, Arro M, Fores O, Manzano D, Ferrer A (2000) Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett 477:170–174
Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RC, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, Haring MA, Giovannoni J, Fernie AR, Klee HJ (2011) Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 23:2738–2753
Dasgupta K, Thilmony R, Stover E, Oliveira ML, Thomson J (2017) Novel R2R3-MYB transcription factors from Prunus americana regulate differential patterns of anthocyanin accumulation in tobacco and citrus. GM Crops Food 8:85–105
D'Auria M, Lorenz R, Racioppi R, Romano VA (2017) Fragrance components of Platanthera bifolia subsp. osca. Nat Prod Res 31:1612–1619
Delorge I, Figueroa CM, Feil R, Lunn JE, Van Dijck P (2015) Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem J 466:283–290
Dudareva N, Murfitt LM, Mann CJ, Gorenstein N, Kolosova N, Kish CM, Bonham C, Wood K (2000) Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12:949–961
Dudareva N, Negre F, Nagegowda DA, Orlova I (2006) Plant volatiles: recent advances and future perspectives. Crit Rev Plant Sci 25:417–440
Feulner M, Schuhwerk F, Dötterl S (2009) Floral scent analysis in Hieracium subgenus Pilosella and its taxonomical implications. Flora 204:495–505
Filion GJ (2015) The signed Kolmogorov-Smirnov test: why it should not be used. Gigascience 4:9
Ginglinger JF et al (2013) Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 25:4640–4657
Goossens J, Mertens J, Goossens A (2017) Role and functioning of bHLH transcription factors in jasmonate signalling. J Exp Bot 68:1333–1347
Grabherr MG et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Guterman I, Masci T, Chen X, Negre F, Pichersky E, Dudareva N, Weiss D, Vainstein A (2006) Generation of phenylpropanoid pathway-derived volatiles in transgenic plants: rose alcohol acetyltransferase produces phenylethyl acetate and benzyl acetate in petunia flowers. Plant Mol Biol 60:555–563
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444:11–25
Hsiao YY, Tsai WC, Kuoh CS, Huang TH, Wang HC, Wu TS, Leu YL, Chen WH, Chen HH (2006) Comparison of transcripts in Phalaenopsis bellina and Phalaenopsis equestris (Orchidaceae) flowers to deduce monoterpene biosynthesis pathway. BMC Plant Biol 6:14
Huizinga DH, Omosegbon O, Omery B, Crowell DN (2008) Isoprenylcysteine methylation and demethylation regulate abscisic acid signaling in Arabidopsis. Plant Cell 20:2714–2728
Jacopini S, Mariani M, de Caraffa VB, Gambotti C, Vincenti S, Desjobert JM, Muselli A, Costa J, Berti L, Maury J (2016) Olive recombinant hydroperoxide lyase, an efficient biocatalyst for synthesis of green leaf volatiles. Appl Biochem Biotechnol 179:671–683
Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Juwattanasomran R, Somta P, Chankaew S, Shimizu T, Wongpornchai S, Kaga A, Srinives P (2011) A SNP in GmBADH2 gene associates with fragrance in vegetable soybean variety “Kaori” and SNAP marker development for the fragrance. Theor Appl Genet 122:533–541
Kanani M, Nazarideljou MJ (2017) Methyl jasmonate and α-aminooxi-β-phenyl propionic acid alter phenylalanine ammonia-lyase enzymatic activity to affect the longevity and floral scent of cut tuberose. Hortic Environ Biotechnol 58:136–143
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Keller Y, Bouvier F, d'Harlingue A, Camara B (1998) Metabolic compartmentation of plastid prenyllipid biosynthesis--evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur J Biochem 251:413–417
Koca N, Karaman Ş (2015) The effects of plant growth regulators and l-phenylalanine on phenolic compounds of sweet basil. Food Chem 166:515–521
Koeduka T, Orlova I, Baiga TJ, Noel JP, Dudareva N, Pichersky E (2009) The lack of floral synthesis and emission of isoeugenol in Petunia axillaris subsp. parodii is due to a mutation in the isoeugenol synthase gene. Plant J 58:961–969
Lan P, Li W, Wang H, Ma W (2010) Characterization, sub-cellular localization and expression profiling of the isoprenylcysteine methylesterase gene family in Arabidopsis thaliana. BMC Plant Biol 10:212
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Lavhale SG, Kalunke RM, Giri AP (2018) Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 248:1063–1078
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Li X, Xu W, Chowdhury M, Jin F (2014) Comparative proteomic analysis of labellum and inner lateral petals in cymbidium ensifolium flowers. Int J Mol Sci 15:19877–19897
Li H, Xiao Q, Zhang C, Du J, Li X, Huang H, Wei B, Li Y, Yu G, Liu H, Hu Y, Liu Y, Zhang J, Huang Y (2017) Identification and characterization of transcription factor ZmEREB94 involved in starch synthesis in maize. J Plant Physiol 216:11–16
Li X, Tang D, Shi Y (2018) Volatile compounds in perianth and corona of Narcissus Pseudonarcissus cultivars. Nat Prod Res:1–4
Liang G, Zhang H, Li X, Ai Q, Yu D (2017) bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J Exp Bot 68:1743–1755
Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199:639–649
Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of Phenylpropanoid metabolism in plants. Mol Plant 8:689–708
Liu F, Xiao Z, Yang L, Chen Q, Shao L, Liu J, Yu Y (2017) PhERF6, interacting with EOBI, negatively regulates fragrance biosynthesis in petunia flowers. New Phytol 215:1490–1502
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
MacMillan J, Takahashi N (1968) Proposed procedure for the allocation of trivial names to the gibberellins. Nature 217:170–171
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Matsui K, Shibata Y, Tateba H, Hatanaka A, Kajiwara T (1997) Changes of lipoxygenase and fatty acid hydroperoxide lyase activities in bell pepper fruits during maturation. Biosci Biotechnol Biochem 61:199–201
Mohd-Hairul AR, Namasivayam P, Cheng Lian GE, Abdullah JO (2010) Terpenoid, benzenoid, and phenylpropanoid compounds in the floral scent of Vanda Mimi palmer. J Plant Biol 53:358–366
Muhlemann JK, Klempien A, Dudareva N (2014) Floral volatiles: from biosynthesis to function. Plant Cell Environ 37:1936–1949
Obiol-Pardo C, Cordero A, Rubio-Martinez J, Imperial S (2010) Homology modeling of mycobacterium tuberculosis 2C-methyl-D-erythritol-4-phosphate cytidylyltransferase, the third enzyme in the MEP pathway for isoprenoid biosynthesis. J Mol Model 16:1061–1073
Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K (2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol 142:1193–1201
Onrubia M, Moyano E, Bonfill M, Palazon J, Goossens A, Cusido RM (2011) The relationship between TXS, DBAT, BAPT and DBTNBT gene expression and taxane production during the development of Taxus baccata plantlets. Plant Sci 181:282–287
Orlova I, Marshall-Colon A, Schnepp J, Wood B, Varbanova M, Fridman E, Blakeslee JJ, Peer WA, Murphy AS, Rhodes D, Pichersky E, Dudareva N (2006) Reduction of Benzenoid synthesis in Petunia flowers reveals multiple pathways to benzoic acid and enhancement in Auxin transport. The Plant Cell Online 18:3458–3475
Pan C, Tian K, Ban Q, Wang L, Sun Q, He Y, Yang Y, Pan Y, Li Y, Jiang J, Jiang C (2017) Genome-wide analysis of the biosynthesis and deactivation of Gibberellin-dioxygenases gene family in Camellia sinensis (L.) O. Kuntze. Genes (Basel) 8
Paul P, Singh SK, Patra B, Sui X, Pattanaik S, Yuan L (2017) A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol 213:1107–1123
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6:3553–3558
Pichersky E, Raguso RA, Lewinsohn E, Croteau R (1994) Floral scent production in Clarkia (Onagraceae) (I. localization and developmental modulation of monoterpene emission and linalool synthase activity). Plant Physiol 106:1533–1540
Pokhilko A, Bou-Torrent J, Pulido P, Rodriguez-Concepcion M, Ebenhoh O (2015) Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol 206:1075–1085
Qu X, Yu B, Liu J, Zhang X, Li G, Zhang D, Li L, Wang X, Wang L, Chen J, Mu W, Pan H, Zhang Y (2014) MADS-box transcription factor SsMADS is involved in regulating growth and virulence in Sclerotinia sclerotiorum. Int J Mol Sci 15:8049–8062
Raguso RA (2016) More lessons from linalool: insights gained from a ubiquitous floral volatile. Curr Opin Plant Biol 32:31–36
Ren Y, Yang J, Lu B, Jiang Y, Chen H, Hong Y, Wu B, Miao Y (2017) Structure of pigment metabolic pathways and their contributions to white tepal color formation of Chinese Narcissus tazetta var chinensis cv Jinzhanyintai. Int J Mol Sci:18
Ruíz-Ramón F, Águila DJ, Egea-Cortines M, Weiss J (2014) Optimization of fragrance extraction: daytime and flower age affect scent emission in simple and double narcissi. Ind Crop Prod 52:671–678
Sakai M, Hirata H, Sayama H, Sekiguchi K, Itano H, Asai T, Dohra H, Hara M, Watanabe N (2014) Production of 2-phenylethanol in roses as the dominant floral scent compound fromL-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Biosci Biotechnol Biochem 71:2408–2419
Sedeek KE, Qi W, Schauer MA, Gupta AK, Poveda L, Xu S, Liu ZJ, Grossniklaus U, Schiestl FP, Schluter PM (2013) Transcriptome and proteome data reveal candidate genes for pollinator attraction in sexually deceptive orchids. PLoS One 8:e64621
Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229:1–13
Song G, Xiao J, Deng C, Zhang X, Hu Y (2007) Use of solid-phase microextraction as a sampling technique for the characterization of volatile compounds emitted from Chinese daffodil flowers. J Anal Chem 62:674–679
Toh C, Mohd-Hairul AR, Ain NM, Namasivayam P, Go R, Abdullah NAP, Abdullah MO, Abdullah JO (2017) Floral micromorphology and transcriptome analyses of a fragrant Vandaceous orchid, Vanda Mimi palmer, for its fragrance production sites. BMC Res Notes 10:554
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511
Van Moerkercke A, Steensma P, Schweizer F, Pollier J, Gariboldi I, Payne R, Vanden Bossche R, Miettinen K, Espoz J, Purnama PC, Kellner F, Seppanen-Laakso T, O'Connor SE, Rischer H, Memelink J, Goossens A (2015) The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc Natl Acad Sci U S A 112:8130–8135
Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339:704–707
Wang Z, Tang J, Hu R, Wu P, Hou XL, Song XM, Xiong AS (2015) Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics 16:17
Wu D, Ji J, Wang G, Guan C, Jin C (2014) LchERF, a novel ethylene-responsive transcription factor from Lycium chinense, confers salt tolerance in transgenic tobacco. Plant Cell Rep 33:2033–2045
Xie DW, Wang XN, Fu LS, Sun J, Zheng W, Li ZF (2015) Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing stress. J Genet 94:55–65
Xu M, Liu C-L, Luo J, Qi Z, Yan Z, Fu Y, Wei S-S, Tang H (2019) Transcriptomic de novo analysis of pitaya (Hylocereus polyrhizus) canker disease caused by Neoscytalidium dimidiatum. BMC Genomics 20
Yamamura C, Mizutani E, Okada K, Nakagawa H, Fukushima S, Tanaka A, Maeda S, Kamakura T, Yamane H, Takatsuji H, Mori M (2015) Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J 84:1100–1113
Yang Z, Endo S, Tanida A, Kai K, Watanabe N (2009) Synergy effect of sodium acetate and glycosidically bound volatiles on the release of volatile compounds from the unscented cut flower (Delphinium elatum L. “Blue Bird”). J Agric Food Chem 57:6396–6401
Yundaeng C, Somta P, Tangphatsornruang S, Chankaew S, Srinives P (2015) A single base substitution in BADH/AMADH is responsible for fragrance in cucumber (Cucumis sativus L.), and development of SNAP markers for the fragrance. Theor Appl Genet 128:1881–1892
Zhao M, Song A, Li P, Chen S, Jiang J, Chen F (2014) A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci Rep 4:6694
Zhu BQ, Cai J, Wang ZQ, Xu XQ, Duan CQ, Pan QH (2014) Identification of a plastid-localized bifunctional nerolidol/linalool synthase in relation to linalool biosynthesis in young grape berries. Int J Mol Sci 15:21992–22010
Acknowledgments
We would like to thank the Charlesworth (https://www.cwauthors.com.cn) for language editing services.
Funding
This work was supported by the Natural Science Foundation of China (NSFC; 30972031) and the Science and Technology Innovation Team of Fujian Academy of Agricultural Sciences (STIT2017-2-11).
Author information
Authors and Affiliations
Contributions
HYS and XM drafted the manuscript together. HYS conceived the study and participated in the design of all experiments. XM performed qRT-PCR and statistical analysis. CXJ participated in the data analysis and discussion about the experiments. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict Interest
Authors declare that they have no conflict interest.
Additional information
Communicated by: Dr. Marcelo C. Dornelas (Associate Editor)
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Table 3
(DOCX 16 kb)
Supplementary Table 4
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
He, Y., Xu, M. & Chen, X. De Novo Transcriptomics Analysis of the Floral Scent of Chinese Narcissus. Tropical Plant Biol. 13, 172–188 (2020). https://doi.org/10.1007/s12042-020-09253-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-020-09253-4