Abstract
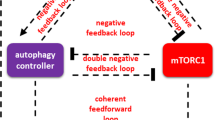
Autophagy is an important cell activity which is the process of formation of autophagosomes, docking with lysosomes and degradation. The intrinsic pathway of apoptosis involves mitochondrial outer membrane permeabilization (MOMP) and cytochrome c release followed by caspase activation. Many molecules, e.g., Ca2+ and mTOR, and different stresses such as endoplasmic reticulum (ER) stress and nutritional stress take part in these two processes. However, the mechanism of how they work together so as to determine cell fate decisions remains to be clarified. Here, we present a computational model for cell fate decisions based on intertwined dynamics with autophagy and apoptosis involving Ca2+, mTOR, and both ER stress and nutritional stress. In agreement with experimental observations, the model predicts that both Ca2+ and the stresses play critical roles in regulating the choice between autophagy and apoptosis in a combinatorial way. The model presented here might be a good candidate for providing the qualitative mechanism of cell fate decisions mediated by Ca2+, mTOR, and two kinds of stress.












Similar content being viewed by others
References
Mizushima N, Autophagy: process and function. Genes Dev. 21(22), 2861–2873 (2007)
Kundu, M., Kosmatka, M., Bardeesy, N., Hurley, R.L., Witters, L.A., DePinho, R.A., Cantley, L.C.: Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112, 1493–1502 (2008)
Loos, B., Engelbrecht, A.M.: Cell death: A dynamic response concept. Autophagy 5(5), 590–603 (2009)
Loos, B., Engelbrecht, A.M., Lockshin, R.A., et al.: The variability of autophagy and cell death susceptibility. Autophagy 9(9), 1270–1285 (2013)
Tyson, J.J., Baumann, W.T., Chen, C., et al.: Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Cell Commun. Signal. 11(7), 523–532 (2011)
Kapuy, O., Vinod, P.K., Mandl, J., et al.: A cellular stress-directed bistable switch controls the crosstalk between autophagy and apoptosis. Mol. BioSyst. 9(2), 296–306 (2013)
Sun, F., Xu, X., Wang, X., et al.: Regulation of autophagy by Ca2 +. Tumor Biol. 37(12), 15467–15476 (2016)
Kapuy, O., Vinod, P.K.: mTOR inhibition increases cell viability via autophagy induction during endoplasmic reticulum stress-an experimental and modeling study. FEBS Open Bio 4, 704–713 (2014)
Dorvash, M., Farahmandnia, M., Tavassoly, I.: A systems biology roadmap to decode mTOR control system in cancer. Interdiscip. Sci. 9, (2019). https://doi.org/10.1007/s12539-019-00347-6
Nikoletopoulou, V., Markaki, M., Palikaras, K., et al.: Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta 1833(12), 3448–3459 (2013)
Bhutia, S.K., Dash, R., Das, S.K., et al.: Mechanism of autophagy to apoptosis switch triggered in prostate cancer cells by antitumor cytokine melanoma differentiation-associated gene 7/interleukin-24. Cancer Res. 70(9), 3667–3676 (2010)
Booth, L.A., Tavallai, S., Hamed, H.A., et al.: The role of cell signalling in the crosstalk between autophagy and apoptosis. Cell. Signal. 26(3), 549–555 (2014)
Kaneko, M., Imaizumi, K., Saito, A., et al.: ER stress and disease: Toward prevention and treatment. Biol. Pharm. Bull. 40(9), 1337–1343 (2017)
Orsolya, K., Papp, D., Tibor, V., et al.: Systems-level feedbacks of NRF2 controlling autophagy upon oxidative stress response. Antioxidants 7(3), 39 (2018)
Liu, B., Oltvai, Z.N., Bayr, H., et al.: Quantitative assessment of cell fate decision between autophagy and apoptosis. Sci. Rep. 7(1), (2017). https://doi.org/10.1038/s41598-017-18001-w
Tavassoly, I., Shajahan, A.N., Parmar, J., et al.: Dynamical modeling of the interaction between autophagy and apoptosis in mammalian cells: a systems pharmacology framework. CPT Pharmacometrics Syst. Pharmacol. 4(4), 263–272 (2013)
Tavassoly, I.: Dynamics of cell fate decision mediated by the interplay of autophagy and apoptosis in cancer cells. Springer International Publishing, Switzerland (2015)
Schleich, K., Lavrik, I.N.: Mathematical modeling of apoptosis. Cell Commun. Signal. 11(1), 44–44 (2013)
Zhang, T., Brazhnik, P., Tyson, J.J.: Computational analysis of dynamical responses to the intrinsic pathway of programmed cell death. Biophys. J. 97(2), 415–434 (2009)
Jin, H., Lei, J.: A mathematical model of cell population dynamics with autophagy response to starvation. Math. Biosci. 258, 1–10 (2014)
Kim, J., Kundu, M., Viollet, B.: AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13(2), 132–141 (2011)
Zhu, W., Chen, Q., Liu, Q., Zhou, Y., Yan, Y., et al.: Study on the interaction between autophagy and apoptosis of MC3T3-E1 at starvation state. J North Sichuan Medical College 2, 206–211(2015) (2015)
Wang, Y., Wang, H., Zuo, L., et al.: Role of autophagy in hepatic stellate cell apoptosis induced by endoplasmic reticulum stress. Acta Univ. Med. Anhui 51(8), 1115–1119 (2016)
Zhang, Y., Ren, S., Liu, Y., et al.: Inhibition of starvation-triggered endoplasmic reticulum stress, autophagy, and apoptosis in ARPE-19 cells by taurine through modulating the expression of calpain-1 and calpain-2. Int. J. Mol. Sci. 18(10), 2146 (2017)
Liu, Y.W., et al.: Bifurcation-based approach reveals synergism and optimal combinatorial perturbation. J. Biol. Phys. 42(3), 399–414 (2016)
Luo, M., Jiao, J., Wang, R.: Screening drug target combinations in disease-related molecular networks. BMC Bioinformatics 20(S7), (2019). https://doi.org/10.1186/s12859-019-2730-8
Funding
This research is supported by the National Natural Science Foundation of China (Grant No. 11971297) and the National Science Foundation of Shanghai (Grant No. 17ZR1410800).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
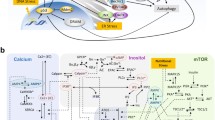
We briefly describe the mathematical models used to study the crosstalk between the autophagy and apoptosis modules. Such a coupled network can be translated into a set of differential equations that describe how each component in the network changes with time. The rate of change of each component is described by ordinary differential equation based on either the law of mass action or Michaelis-Menten kinetics [5, 7, 14, 15].
1.1 Apoptosis
1.2 Autophagy
1.3 The coupled model
Rights and permissions
About this article
Cite this article
Ge, Z., Wang, R. Fate decisions mediated by crosstalk of autophagy and apoptosis in mammalian cells. J Biol Phys 46, 133–149 (2020). https://doi.org/10.1007/s10867-020-09542-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10867-020-09542-9