Abstract
Background
Grape is an economically valuable fruit around the world. However, some cultivars are prone to fruit cracking during ripening, leading to severe losses.
Objective
We aimed to find important metabolisms related to fruit cracking during ripening process.
Methods
RNA-Sequence and analysis was applied to the pericarp of cracking-susceptible ‘Xiang Fei’ at 1 (W1), 2 (W2) and 3 weeks (W3) after veraison on Illumina HiSeq xten;
Results
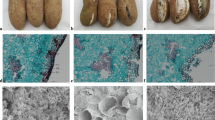
Compared with W1, the berry cracking rate increased significantly in W2 and W3. Through transcriptomic analysis, a total of 22,609 genes were expressed in the grape pericarp, among which 805 and 2758 genes were significantly differentially regulated in W1-vs.-W2 and W1-vs.-W3 comparison, respectively. Besides, 304 and 354 genes were up- and down-regulated in both comparisons. The significantly enriched GO terms of both W1–W2 and W1–W3 are related to cell wall and wax biosynthesis. And lipid metabolism, which are involved in the top 20 enriched KEGG pathways of both comparisons, was related to wax biosynthesis. Further, GO enrichment analysis of differentially expressed genes (DEGs) with same regulatory changes also indicated that the continuously up-regulated DEGs are significantly enriched in cell wall component biosynthesis and hydrolase.
Conclusion
These findings suggested that genes related to cell wall metabolism and cuticle biosynthesis may play important roles in regulating grape berry cracking. Our results provide a reference for further studies on the molecular mechanism underlying fruit cracking.








Similar content being viewed by others
References
Agustí M, Mesejo C (2002) Citrus fruit quality. Physiology basis and techniques of improvement. Agrociencia 6:1–16
Almela V, Zaragoza S, Primo-Millo E, Agusti M (1994) Hormonal control of splitting in ‘Nova’ mandarin fruit. J Pomol Hortic Sci 69:969–973. https://doi.org/10.1080/00221589.1994.11516534
Becker T, Knoche M (2012) Deposition, strain, and microcracking of the cuticle in developing “Riesling” grape berries. Vitis 51:1–6
Brüggenwirth M, Knoche M (2016) Mechanical properties of skins of sweet cherry fruit of differing susceptibilities to cracking. J Am Soc Hortic Sci 141:162–168. https://doi.org/10.21273/JASHS.141.2.162
Brüggenwirth M, Knoche M (2017) Cell wall swelling, fracture mode, and the mechanical properties of cherry fruit skins are closely related. Planta 245:765–777. https://doi.org/10.1007/s00425-016-2639-7
Brummell DA (2006) Cell wall disassembly in ripening fruit. Funct Plant Biol 33:103–119. https://doi.org/10.1071/FP05234
Brummell DA, Harpster MH (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol 47:311–339. https://doi.org/10.1023/a:1010656104304
Cao K, Wang LR, Zhu GR, Fang WC, Chen CW, Feng YB (2009) Comparison of anatomical fruit structure in different types of peach during fruit development. J Fruit Sci 26:440–444. https://doi.org/10.13925/j.cnki.gsxb.2009.04.032
Cao YB, Li CJ, Sun F, Zhang LY (2014) Comparison of the endogenous hormones content and the activities of enzymes related to cell-wall metabolism between jujube cultivars susceptible and resistant to fruit cracking. Acta Hortic Sin 41:139–148. https://doi.org/10.3969/j.issn.0513-353X.2014.01.016
Capel C, Yuste-Lisbona FJ, López-Casado G, Angosto T, Cuartero J, Lozano R, Capel J (2017) Multi-environment QTL mapping reveals genetic architecture of fruit cracking in a tomato RIL Solanum lycopersicum × S. pimpinellifolium population. Theor Appl Genet 130:213–222. https://doi.org/10.1007/s00122-016-2809-9
Chen K, Li EC, Xu ZX, Li TY, Xu C, Qin JG, Chen LQ (2015) Comparative transcriptome analysis in the hepatopancreas tissue of pacific white shrimp litopenaeus vannamei fed different lipid sources at low salinity. PLoS One 10:e0144889. https://doi.org/10.1371/journal.pone.0144889
Choi H, Son I, Kim D (2010) Effects of calcium concentrations of coating bag on pericarp structure and berry cracking in ‘Kyoho’ grape (Vitis sp.). Korean J Hortic Sci Technol 28:561–566
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Considine JA, Knox RB (1979) Development and histochemistry of the cells, cell walls, and cuticle of the dermal system of fruit of the grape, Vitis vinifera L. Protoplasma 99:347–365. https://doi.org/10.1007/bf01275807
Considine J, Kriedemann PE (1972) Fruit splitting in grapes: determination of the critical turgor pressure. Aust J Agric Res 23:17–23. https://doi.org/10.1071/AR9720017
Coombe BG (1992) Research on development and ripening of the grape berry. Am J Enol Vitic 43:101
Cristián B, Héctor A, Joselyn R, Jessica C, Carlos RF (2014) Transcriptional analysis of cell wall and cuticle related genes during fruit development of two sweet cherry cultivars with contrasting levels of cracking tolerance. Chil J Agric Res 74:162–169. https://doi.org/10.4067/S0718-58392014000200006
Dominguez E, Cuartero J, Heredia A (2011) An overview on plant cuticle biomechanics. Plant Sci 181:77–84. https://doi.org/10.1016/j.plantsci.2011.04.016
Domínguez E, Fernández MD, Hernández JCL, Parra JP, España L, Heredia A, Cuartero J (2012) Tomato fruit continues growing while ripening, affecting cuticle properties and cracking. Physiol Plant 146:473–486. https://doi.org/10.1111/j.1399-3054.2012.01647.x
Garcia-Luis A, Duarte AMM, Porras I, García-Lidón A, Guardiola JL (1994) Fruit splitting in ‘Nova’ hybrid mandarin in relation to the anatomy of the fruit and fruit set treatments. Sci Hortic 57:215–231. https://doi.org/10.1016/0304-4238(94)90142-2
Gibert C, Chadœuf J, Vercambre G, Génard M, Lescourret F (2007) Cuticular cracking on nectarine fruit surface: spatial distribution and development in relation to irrigation and thinning. J Am Soc Hortic Sci 132:583–591. https://doi.org/10.21273/JASHS.132.5.583
Ginzberg I, Fogelman E, Rosenthal L, Stern RA (2014) Maintenance of high epidermal cell density and reduced calyx-end cracking in developing ‘Pink Lady’ apples treated with a combination of cytokinin 6-benzyladenine and gibberellins A4 + A7. Sci Hortic 165:324–330. https://doi.org/10.1016/j.scienta.2013.11.020
Huang XM, Li JG, Wang HC, Huang HB, Gao FF (2001) The relationship between fruit cracking and calcium in litchi pericarp. Acta Hortic 558:209–215. https://doi.org/10.17660/actahortic.2001.558.29
Huang XM, Yuan WQ, Wang HC, Li JG, Huang HB, Shi L, Yin JH (2004) Linking cracking resistance and fruit desiccation rate to pericarp structure in litchi (Litchi chinensis Sonn.). J Hortic Sci Biotechnol 79:897–905. https://doi.org/10.1080/14620316.2004.11511863
Inchang S, Seonkyu K, Haghyun K, Gilhah K (2007) Physiological and histological characteristics of berry cracking in grapes (Vitis spp.). Hortic Environ Biotechnol 48:678–685. https://doi.org/10.1177/030098588001700604
Jiang FL, Lopez A, Jeon S, de Freitas ST, Yu QH, Wu Z, Labavitch JM, Tian SK, Powell ALT, Mitcham E (2019) Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking. Hortic Res 6:17. https://doi.org/10.1038/s41438-018-0105-3
John MA, Dey PM (1986) Postharvest changes in fruit cell wall. Adv Food Res 30:139–193. https://doi.org/10.1016/S0065-2628(08)60349-3
Kasai S, Hayama H, Kashimura Y, Kudo S, Osanai Y (2008) Relationship between fruit cracking and expression of the expansin gene MdEXPA3 in ‘Fuji’ apples (Malusdomestica Borkh.). Sci Hortic 116:194–198. https://doi.org/10.1016/j.scienta.2007.12.002
Khadivi-Khub A (2014) Physiological and genetic factors influencing fruit cracking. Acta Physiol Plant 37:1718. https://doi.org/10.1007/s11738-014-1718-2
Lane WD, Meheriuk M, McKenzie DL (2000) Fruit cracking of a susceptible, an intermediate, and a resistant sweet cherry cultivar. HortScience 35:239–242. https://doi.org/10.21273/HORTSCI.35.2.239
Lang A, Düring H (1990) Grape berry splitting and some mechanical properties of the skin. Vitis 29:61–70. https://doi.org/10.1016/0304-4238(90)90030-I
Leng F, Lin Q, Wu D, Wang S, Wang D, Sun C (2016) Comparative transcriptomic analysis of grape berry in response to root restriction during developmental stages. Molecules 21:1431. https://doi.org/10.3390/molecules21111431
Letaief H, Rolle L, Zeppa G, Gerbi V (2008) Assessment of grape skin hardness by a puncture test. J Sci Food Agric 88:1567–1575. https://doi.org/10.1002/jsfa.3252
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323. https://doi.org/10.1186/1471-2105-12-323
Li JG, Huang XM, Huang HB (2003) Comparison of the activities of enzymes related to cell-Wa metabolism in pericarp between litchi cultivars susceptible and resistant to fruit cracking. J Plant Physiol Mol Biol 29:141–146. https://doi.org/10.3321/j.issn:1671-3877.2003.02.010
Li WC, Wu JY, Zhang HN, Shi SY, Liu LQ, Shu B, Liang QZ, Xie JH, Wei YZ (2014) De novo assembly and characterization of pericarp transcriptome and identification of candidate genes mediating fruit cracking in Litchi chinensis Sonn. Int J Mol Sci 15:17667–17685. https://doi.org/10.3390/ijms151017667
Li CX, Xu ZG, Dong RQ, Chang SX, Wang LZ, Khalil-Ur-Rehman M, Tao JM (2017) An RNA-Seq analysis of grape plantlets grown in vitro reveals different responses to blue, green, red LED light, and white fluorescent light. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00078
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lu PL, Lin CH (2011) Physiology of fruit cracking in wax apple (Syzygium samarangense). Bot Orient J Plant Sci 8:70–76. https://doi.org/10.3126/botor.v8i0.5954
Meynhardt JT (1964) A histological study of berry-splitting in some grape cultivars. S Afr J Agric Sci 7:707–716
Nunan KJ, Davies C, Robinson SP, Fincher GB (2001) Expression patterns of cell wall-modifying enzymes during grape berry development. Planta 214:257–264. https://doi.org/10.1007/s004250100609
Opara LU, Hodson AJ, Studman CJ (2015) Stem-end splitting and internal ring-cracking of ‘Gala’ apples as influenced by orchard management practices. J Hortic Sci Biotechnol 75:465–469. https://doi.org/10.1080/14620316.2000.11511270
Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284:1976. https://doi.org/10.1126/science.284.5422.1976
Peschel S, Knoche M (2005) Characterization of microcracks in the cuticle of developing sweet cherry fruit. J Am Soc Hortic Sci 130:487–495
Ramteke SD, Urkude V, Parhe SD, Bhagwat SR (2017) Berry cracking; its causes and remedies in grapes—a review. Trends Biosci 10:549–556
Riederer M, Schreiber L (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52:2023–2032. https://doi.org/10.1093/jexbot/52.363.2023
Rios JC, Robledo F, Schreiber L, Zeisler V, Lang E, Carrasco B, Silva H (2015) Association between the concentration of n-alkanes and tolerance to cracking in commercial varieties of sweet cherry fruits. Sci Hortic 197:57–65. https://doi.org/10.1016/j.scienta.2015.10.037
Sahu P, Sharma N, Sharma DP (2013) Effect of in situ moisture conservation, forchlorfenuron and boron on growth, fruit cracking and yield of pomegranate cv. Kandhari under rainfed conditions of Himachal Pradesh. Indian J Hortic 70:501–505
Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59:683–707. https://doi.org/10.1146/annurev.arplant.59.103006.093219
Schuch W, Kanczler JM, Robertson D, Hobson G, Tucker G, Grierson D, Bright S, Bird C (1991) Fruit-quality characteristics of transgenic tomato fruit with altered polygalacturonase activity. HortScience 26:1517–1520
Sekse L (1995) Fruit cracking in sweet cherries (Prunus avium L.). Some physiological aspects—a mini review. Sci Hortic 63:135–141. https://doi.org/10.1016/0304-4238(95)00806-5
Shen YH, Lu BG, Feng L, Yang FY, Geng JJ, Ming R, Chen XJ (2017) Isolation of ripening-related genes from ethylene/1-MCP treated papaya through RNA-seq. BMC Genom 18:671. https://doi.org/10.1186/s12864-017-4072-0
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800–e21800. https://doi.org/10.1371/journal.pone.0021800
Varet H, Brillet-Guéguen L, Coppée J-Y, Dillies M-A (2016) SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One 11:e0157022. https://doi.org/10.1371/journal.pone.0157022
Vicens A, Fournand D, Williams P, Sidhoum L, Moutounet M, Doco T (2009) Changes in Polysaccharide and protein composition of cell walls in grape berry skin (Cv. Shiraz) during ripening and over-ripening. J Agric Food Chem 57:2955–2960. https://doi.org/10.1021/jf803416w
Wang XX, Fan XC, Li A, Zhang CB, Fang JG, Liu CH, Shangguan LF (2016) Investigation and analysis on cracking trait in grape berry. Acta Hortic Sin. https://doi.org/10.16420/j.issn.0513-353x.2016-0122
Wang X, Lin LJ, Tang Y, Xia H, Zhang XC, Yue ML, Qiu X, Xu K, Wang ZH (2018) Transcriptomic insights into citrus segment membrane’s cell wall components relating to fruit sensory texture. BMC Genom 19:280. https://doi.org/10.1186/s12864-018-4669-y
Wang JG, Gao XM, Ma ZL, Chen J, Liu YN (2019) Analysis of the molecular basis of fruit cracking susceptibility in Litchi chinensis cv. Baitangying by transcriptome and quantitative proteome profiling. J Plant Physiol 234:106–116. https://doi.org/10.1016/j.jplph.2019.01.014
Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, Kong L, Gao G, Li CY, Wei LP (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. https://doi.org/10.1093/nar/gkr483
Yamamura H, Naito R (1985) Susceptibility to berry splitting in several grape cultivars. J Jpn Soc Hortic Sci 53:390–395
Yang ZE, Wu Z, Zhang C, Hu EM, Zhou R, Jiang FL (2016) The composition of pericarp, cell aging, and changes in water absorption in two tomato genotypes: mechanism, factors, and potential role in fruit cracking. Acta Physiol Plant 38:215. https://doi.org/10.1007/s11738-016-2228-1
Yilmaz C, Ozgüven AI (2006) Hormone physiology of preharvest fruit cracking in pomegranate (Punica granatum L.). Acta Hortic 727:545–550. https://doi.org/10.17660/actahortic.2006.727.67
Zoffoli JP, Latorre BA, Naranjo P (2008) Hairline, a postharvest cracking disorder in table grapes induced by sulfur dioxide. Postharvest Biol Technol 47:90–97. https://doi.org/10.1016/j.postharvbio.2007.06.013
Zsófi Z, Villangó S, Pálfi Z, Tóth E, Bálo B (2014) Texture characteristics of the grape berry skin and seed (Vitis vinifera L. cv. Kékfrankos) under postveraison water deficit. Sci Hortic 172:176–182. https://doi.org/10.1016/j.scienta.2014.04.008
Acknowledgements
We thank Jennifer Smith, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This research was funded by National Technology System for Grape Industry, Grant number CARS-29-ZP-9.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, M., Yu, J., Zhao, M. et al. Transcriptome analysis of metabolisms related to fruit cracking during ripening of a cracking-susceptible grape berry cv. Xiangfei (Vitis vinifera L.). Genes Genom 42, 639–650 (2020). https://doi.org/10.1007/s13258-020-00930-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-020-00930-y