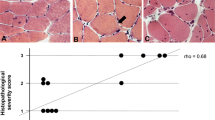
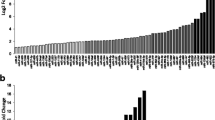
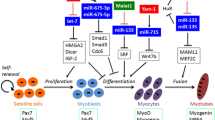
Abstract
Noncoding RNAs (ncRNAs), such as miRNAs and long noncoding RNAs, are key regulators of gene expression at the post-transcriptional level and represent promising therapeutic targets and biomarkers for several human diseases, including Duchenne and Becker muscular dystrophies (DMD/BMD). A role for ncRNAs in the pathogenesis of muscular dystrophies has been suggested, even if it is still incompletely understood. Here, we discuss current progress leading towards the clinical utility of ncRNAs for DMD/BMD. Long and short noncoding RNAs are differentially expressed in DMD/BMD and have a mechanism of action via targeting mRNAs. A subset of muscle-enriched miRNAs, the so-called myomiRs (miR-1, miR-133, and miR-206), are increased in the serum of patients with DMD and in dystrophin-defective animal models. Interestingly, myomiRs might be used as biomarkers, given that their levels can be corrected after dystrophin restoration in dystrophic mice. Remarkably, further evidence demonstrates that ncRNAs also play a role in dystrophin expression; thus, their modulations might represent a potential therapeutic strategy with the aim of upregulating the dystrophin protein in combination with other oligonucleotides/gene therapy approaches.

Similar content being viewed by others
References
Emery AE (1991) Population frequencies of inherited neuromuscular diseases—a world survey. Neuromuscul Disord NMD 1:19–29
Mah JK, Korngut L, Dykeman J et al (2014) A systematic review and meta-analysis on the epidemiology of Duchenne and Becker muscular dystrophy. Neuromuscul Disord NMD 24:482–491. https://doi.org/10.1016/j.nmd.2014.03.008
Monaco AP, Bertelson CJ, Liechti-Gallati S et al (1988) An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2:90–95
Gao QQ, McNally EM (2015) The dystrophin complex: structure, function, and implications for therapy. Compr Physiol 5:1223–1239. https://doi.org/10.1002/cphy.c140048
Petrof BJ, Shrager JB, Stedman HH et al (1993) Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci 90:3710–3714. https://doi.org/10.1073/pnas.90.8.3710
Fairclough RJ, Perkins KJ, Davies KE (2012) Pharmacologically targeting the primary defect and downstream pathology in Duchenne muscular dystrophy. Curr Gene Ther 12:206–244. https://doi.org/10.2174/156652312800840595
Boldrin L, Zammit PS, Morgan JE (2015) Satellite cells from dystrophic muscle retain regenerative capacity. Stem Cell Res 14:20–29. https://doi.org/10.1016/j.scr.2014.10.007
Cohn RD, Campbell KP (2000) Molecular basis of muscular dystrophies. Muscle Nerve 23:1456–1471. https://doi.org/10.1002/1097-4598(200010)23:10%3c1456:AID-MUS2%3e3.0.CO;2-T
Nichols B, Takeda S, Yokota T (2015) Nonmechanical roles of dystrophin and associated proteins in exercise, neuromuscular junctions, and brains. Brain Sci 5:275–298. https://doi.org/10.3390/brainsci5030275
Constantin B (2014) Dystrophin complex functions as a scaffold for signalling proteins. Biochim Biophys Acta BBA Biomembr 1838:635–642. https://doi.org/10.1016/j.bbamem.2013.08.023
Darras BT, Urion DK, Ghosh PS (1993) Dystrophinopathies. In: Adam MP, Ardinger HH, Pagon RA, et al (eds) GeneReviews®. University of Washington, Seattle, Seattle
Sussman M (2002) Duchenne muscular dystrophy. J Am Acad Orthop Surg 10:138–151
Brandsema JF, Darras BT (2015) Dystrophinopathies. Semin Neurol 35:369–384. https://doi.org/10.1055/s-0035-1558982
Nigro G, Comi LI, Politano L, Bain RJ (1990) The incidence and evolution of cardiomyopathy in Duchenne muscular dystrophy. Int J Cardiol 26:271–277
Magri F, Govoni A, D’Angelo MG et al (2011) Genotype and phenotype characterization in a large dystrophinopathic cohort with extended follow-up. J Neurol 258:1610–1623. https://doi.org/10.1007/s00415-011-5979-z
Yazaki M, Yoshida K, Nakamura A et al (1999) Clinical characteristics of aged Becker muscular dystrophy patients with onset after 30 years. Eur Neurol 42:145–149. https://doi.org/10.1159/000008089
Nigro G, Comi LI, Politano L et al (1995) Evaluation of the cardiomyopathy in Becker muscular dystrophy. Muscle Nerve 18:283–291. https://doi.org/10.1002/mus.880180304
Govoni A, Magri F, Brajkovic S et al (2013) Ongoing therapeutic trials and outcome measures for Duchenne muscular dystrophy. Cell Mol Life Sci CMLS 70:4585–4602. https://doi.org/10.1007/s00018-013-1396-z
Mendell JR, Goemans N, Lowes LP et al (2016) Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann Neurol 79:257–271. https://doi.org/10.1002/ana.24555
Lim KRQ, Maruyama R, Yokota T (2017) Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des Devel Ther 11:533–545. https://doi.org/10.2147/DDDT.S97635
Heo Y-A (2020) Golodirsen: first approval. Drugs 80:329–333. https://doi.org/10.1007/s40265-020-01267-2
Bushby K, Finkel R, Wong B et al (2014) Ataluren treatment of patients with nonsense mutation dystrophinopathy: ataluren for dystrophinopathy. Muscle Nerve 50:477–487. https://doi.org/10.1002/mus.24332
Duan D (2018) Systemic AAV micro-dystrophin gene therapy for duchenne muscular dystrophy. Mol Ther J Am Soc Gene Ther 26:2337–2356. https://doi.org/10.1016/j.ymthe.2018.07.011
Matre PR, Mu X, Wu J et al (2019) CRISPR/Cas9-based dystrophin restoration reveals a novel role for dystrophin in bioenergetics and stress resistance of muscle progenitors. Stem Cells Dayt Ohio 37:1615–1628. https://doi.org/10.1002/stem.3094
Fiorillo AA, Heier CR, Novak JS et al (2015) TNF-α-induced microRNAs control dystrophin expression in becker muscular dystrophy. Cell Rep 12:1678–1690. https://doi.org/10.1016/j.celrep.2015.07.066
Verhaart IEC, Aartsma-Rus A (2019) Therapeutic developments for Duchenne muscular dystrophy. Nat Rev Neurol 15:373–386. https://doi.org/10.1038/s41582-019-0203-3
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15 Spec No 1:R17–R29. https://doi.org/10.1093/hmg/ddl046
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10:925–933. https://doi.org/10.4161/rna.24604
Hombach S, Kretz M (2016) Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol 937:3–17. https://doi.org/10.1007/978-3-319-42059-2_1
Trzybulska D, Vergadi E (2018) MiRNA and other non-coding RNAs as promising diagnostic markers. EJIFCC 29(3):221–226
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531. https://doi.org/10.1038/nrg1379
Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M (2014) Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genom. https://doi.org/10.1155/2014/970607
Bu D, Luo H, Jiao F et al (2015) Evolutionary annotation of conserved long non-coding RNAs in major mammalian species. Sci China Life Sci 58:787–798. https://doi.org/10.1007/s11427-015-4881-9
Geisler S, Coller J (2013) RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14:699–712. https://doi.org/10.1038/nrm3679
Zhang X, Wang W, Zhu W et al (2019) Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. https://doi.org/10.3390/ijms20225573
Bak RO, Mikkelsen JG (2014) miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA 5:317–333. https://doi.org/10.1002/wrna.1213
Paraskevopoulou MD, Hatzigeorgiou AG (2016) Analyzing MiRNA–LncRNA interactions. In: Feng Y, Zhang L (eds) Long non-coding RNAs: methods and protocols. Springer, New York, pp 271–286
Bovolenta M, Erriquez D, Valli E et al (2012) The DMD locus harbours multiple long non-coding RNAs which orchestrate and control transcription of muscle dystrophin mRNA isoforms. PLoS One 7:e45328. https://doi.org/10.1371/journal.pone.0045328
Greco S, De Simone M, Colussi C et al (2009) Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J 23:3335–3346. https://doi.org/10.1096/fj.08-128579
Fiorillo AA, Tully CB, Damsker JM et al (2018) Muscle miRNAome shows suppression of chronic inflammatory miRNAs with both prednisone and vamorolone. Physiol Genom 50:735–745. https://doi.org/10.1152/physiolgenomics.00134.2017
Liu DZ, Stamova B, Hu S et al (2015) MicroRNA and mRNA expression changes in steroid naïve and steroid treated DMD patients. J Neuromuscul Dis 2:387–396. https://doi.org/10.3233/JND-150076
Zaharieva IT, Calissano M, Scoto M et al (2013) Dystromirs as serum biomarkers for monitoring the disease severity in duchenne muscular dystrophy. PLoS One 8:e80263. https://doi.org/10.1371/journal.pone.0080263
Taganov KD, Boldin MP, Chang K-J, Baltimore D (2006) NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103:12481–12486. https://doi.org/10.1073/pnas.0605298103
Wei W, He H-B, Zhang W-Y et al (2013) miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis 4:e668. https://doi.org/10.1038/cddis.2013.184
Wang H, Garzon R, Sun H et al (2008) NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14:369–381. https://doi.org/10.1016/j.ccr.2008.10.006
Wang L, Zhou L, Jiang P et al (2012) Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol Ther 20:1222–1233. https://doi.org/10.1038/mt.2012.35
Chen J-F, Mandel EM, Thomson JM et al (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38:228–233. https://doi.org/10.1038/ng1725
Nguyen-Tran D-H, Hait NC, Sperber H et al (2014) Molecular mechanism of sphingosine-1-phosphate action in Duchenne muscular dystrophy. Dis Model Mech 7:41–54. https://doi.org/10.1242/dmm.013631
Ardite E, Perdiguero E, Vidal B et al (2012) PAI-1–regulated miR-21 defines a novel age-associated fibrogenic pathway in muscular dystrophy. J Cell Biol 196:163–175. https://doi.org/10.1083/jcb.201105013
Zanotti S, Gibertini S, Curcio M et al (2015) Opposing roles of miR-21 and miR-29 in the progression of fibrosis in Duchenne muscular dystrophy. Biochim Biophys Acta BBA Mol Basis Dis 1852:1451–1464. https://doi.org/10.1016/j.bbadis.2015.04.013
Zanotti S, Gibertini S, Blasevich F et al (2018) Exosomes and exosomal miRNAs from muscle-derived fibroblasts promote skeletal muscle fibrosis. Matrix Biol 74:77–100. https://doi.org/10.1016/j.matbio.2018.07.003
Giordani L, Sandoná M, Rotini A et al (2014) Muscle-specific microRNAs as biomarkers of Duchenne Muscular Dystrophy progression and response to therapies. Rare Dis 2:e974969. https://doi.org/10.4161/21675511.2014.974969
Saccone V, Consalvi S, Giordani L et al (2014) HDAC-regulated myomiRs control BAF60 variant exchange and direct the functional phenotype of fibro-adipogenic progenitors in dystrophic muscles. Genes Dev 28:841–857. https://doi.org/10.1101/gad.234468.113
Cacchiarelli D, Martone J, Girardi E et al (2010) MicroRNAs involved in molecular circuitries relevant for the duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab 12:341–351. https://doi.org/10.1016/j.cmet.2010.07.008
Guilbaud M, Gentil C, Peccate C et al (2018) miR-708-5p and miR-34c-5p are involved in nNOS regulation in dystrophic context. Skelet Muscle. https://doi.org/10.1186/s13395-018-0161-2
Cordani N, Pisa V, Pozzi L et al (2014) Nitric oxide controls fat deposition in dystrophic skeletal muscle by regulating fibro-adipogenic precursor differentiation. Stem Cells Dayt Ohio 32:874–885. https://doi.org/10.1002/stem.1587
De Arcangelis V, Serra F, Cogoni C et al (2010) β1-syntrophin modulation by miR-222 in mdx mice. PLoS One. https://doi.org/10.1371/journal.pone.0012098
Robriquet F, Babarit C, Larcher T et al (2016) Identification in GRMD dog muscle of critical miRNAs involved in pathophysiology and effects associated with MuStem cell transplantation. BMC Musculoskelet Disord. https://doi.org/10.1186/s12891-016-1060-5
Morgan JE, Partridge TA (2003) Muscle satellite cells. Int J Biochem Cell Biol 35:1151–1156. https://doi.org/10.1016/s1357-2725(03)00042-6
von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA (2013) Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci USA 110:16474–16479. https://doi.org/10.1073/pnas.1307680110
Bentzinger CF, Wang YX, Rudnicki MA (2012) Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a008342
Buckingham M, Rigby PWJ (2014) Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell 28:225–238. https://doi.org/10.1016/j.devcel.2013.12.020
Zammit PS (2017) Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol 72:19–32. https://doi.org/10.1016/j.semcdb.2017.11.011
Asfour HA, Allouh MZ, Said RS (2018) Myogenic regulatory factors: the orchestrators of myogenesis after 30 years of discovery. Exp Biol Med Maywood NJ 243:118–128. https://doi.org/10.1177/1535370217749494
Ballarino M, Morlando M, Fatica A, Bozzoni I (2016) Non-coding RNAs in muscle differentiation and musculoskeletal disease. J Clin Invest 126:2021–2030. https://doi.org/10.1172/JCI84419
Nie M, Deng Z-L, Liu J, Wang D-Z (2015) Noncoding RNAs, emerging regulators of skeletal muscle development and diseases. Biomed Res Int 2015:676575. https://doi.org/10.1155/2015/676575
Hagan M, Zhou M, Ashraf M, et al (2017) Long noncoding RNAs and their roles in skeletal muscle fate determination. Non-Coding RNA Investig. https://doi.org/10.21037/ncri.2017.12.01
Cacchiarelli D, Incitti T, Martone J et al (2011) miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep 12:136–141. https://doi.org/10.1038/embor.2010.208
Hindi SM, Kumar A (2016) TRAF6 regulates satellite stem cell self-renewal and function during regenerative myogenesis. J Clin Invest 126:151–168. https://doi.org/10.1172/JCI81655
Ling Y-H, Sui M-H, Zheng Q et al (2018) miR-27b regulates myogenic proliferation and differentiation by targeting Pax3 in goat. Sci Rep 8:1–12. https://doi.org/10.1038/s41598-018-22262-4
Kuang W, Tan J, Duan Y et al (2009) Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem Biophys Res Commun 378:259–263. https://doi.org/10.1016/j.bbrc.2008.11.041
Khanna N, Ge Y, Chen J (2014) MicroRNA-146b promotes myogenic differentiation and modulates multiple gene targets in muscle cells. PLoS One 9:e100657. https://doi.org/10.1371/journal.pone.0100657
Kim HK, Lee YS, Sivaprasad U et al (2006) Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174:677–687. https://doi.org/10.1083/jcb.200603008
Wang XH, Hu Z, Klein JD et al (2011) Decreased miR-29 suppresses myogenesis in CKD. J Am Soc Nephrol JASN 22:2068–2076. https://doi.org/10.1681/ASN.2010121278
Naguibneva I, Ameyar-Zazoua M, Polesskaya A et al (2006) The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol 8:278–284. https://doi.org/10.1038/ncb1373
Wang L, Chen X, Zheng Y et al (2012) MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp Cell Res 318:2324–2334. https://doi.org/10.1016/j.yexcr.2012.06.018
Antoniou A, Mastroyiannopoulos NP, Uney JB, Phylactou LA (2014) miR-186 inhibits muscle cell differentiation through myogenin regulation. J Biol Chem 289:3923–3935. https://doi.org/10.1074/jbc.M113.507343
Sun Q, Zhang Y, Yang G et al (2008) Transforming growth factor-β-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res 36:2690–2699. https://doi.org/10.1093/nar/gkn032
Cardinali B, Castellani L, Fasanaro P et al (2009) Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One 4:e7607. https://doi.org/10.1371/journal.pone.0007607
Eisenberg I, Eran A, Nishino I et al (2007) Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci 104:17016–17021. https://doi.org/10.1073/pnas.0708115104
Crist CG, Montarras D, Buckingham M (2012) Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 11:118–126. https://doi.org/10.1016/j.stem.2012.03.011
Jia L, Li Y-F, Wu G-F et al (2013) MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int J Mol Sci 15:296–308. https://doi.org/10.3390/ijms15010296
Mousavi K, Zare H, Dell’orso S et al (2013) eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51:606–617. https://doi.org/10.1016/j.molcel.2013.07.022
Hubé F, Velasco G, Rollin J et al (2011) Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res 39:513–525. https://doi.org/10.1093/nar/gkq833
Borensztein M, Monnier P, Court F et al (2013) Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Dev Camb Engl 140:1231–1239. https://doi.org/10.1242/dev.084665
Dey BK, Pfeifer K, Dutta A (2014) The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 28:491–501. https://doi.org/10.1101/gad.234419.113
Zhang K, Sha J, Harter ML (2010) Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol 188:39–48. https://doi.org/10.1083/jcb.200904144
Guo Y, Wang J, Zhu M et al (2017) Identification of MyoD-responsive transcripts reveals a novel long non-coding RNA (lncRNA-AK143003) that negatively regulates myoblast differentiation. Sci Rep 7:2828. https://doi.org/10.1038/s41598-017-03071-7
Wang L, Zhao Y, Bao X et al (2015) LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res 25:335–350. https://doi.org/10.1038/cr.2015.21
Lu L, Sun K, Chen X et al (2013) Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J 32:2575–2588. https://doi.org/10.1038/emboj.2013.182
Chen X, He L, Zhao Y et al (2017) Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov 3:17002. https://doi.org/10.1038/celldisc.2017.2
Zhou L, Sun K, Zhao Y et al (2015) Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun 6:10026. https://doi.org/10.1038/ncomms10026
Yu X, Zhang Y, Li T et al (2017) Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat Commun 8:14016. https://doi.org/10.1038/ncomms14016
Mueller AC, Cichewicz MA, Dey BK et al (2015) MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol Cell Biol 35:498–513. https://doi.org/10.1128/MCB.01079-14
Wang G, Wang Y, Xiong Y et al (2016) Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep 6:21865. https://doi.org/10.1038/srep21865
Zhu M, Liu J, Xiao J et al (2017) Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat Commun 8:14718. https://doi.org/10.1038/ncomms14718
Zhang Z, Li J, Guan D et al (2018) A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J Cachexia Sarcopenia Muscle 9:613–626. https://doi.org/10.1002/jcsm.12281
Cesana M, Cacchiarelli D, Legnini I et al (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369. https://doi.org/10.1016/j.cell.2011.09.028
Ballarino M, Cazzella V, D’Andrea D et al (2015) novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol Cell Biol 35:728–736. https://doi.org/10.1128/MCB.01394-14
Nakasa T, Ishikawa M, Shi M et al (2010) Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med 14:2495–2505. https://doi.org/10.1111/j.1582-4934.2009.00898.x
Butchart LC, Terrill JR, Rossetti G et al (2018) Expression patterns of regulatory RNAs, including lncRNAs and tRNAs, during postnatal growth of normal and dystrophic (mdx) mouse muscles, and their response to taurine treatment. Int J Biochem Cell Biol 99:52–63. https://doi.org/10.1016/j.biocel.2018.03.016
Nghiem PP, Hoffman EP, Mittal P et al (2013) Sparing of the dystrophin-deficient cranial sartorius muscle is associated with classical and novel hypertrophy pathways in GRMD dogs. Am J Pathol 183:1411–1424. https://doi.org/10.1016/j.ajpath.2013.07.013
Alexander MS, Casar J, Motohashi N et al (2011) Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet Muscle 1:27. https://doi.org/10.1186/2044-5040-1-27
Alexander MS, Casar JC, Motohashi N et al (2014) MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J Clin Invest 124:2651–2667. https://doi.org/10.1172/JCI73579
Alexander MS, Kawahara G, Motohashi N et al (2013) MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation and myogenic differentiation. Cell Death Differ 20:1194–1208. https://doi.org/10.1038/cdd.2013.62
Liu J, Liang X, Zhou D et al (2016) Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol Med 8:1212–1228. https://doi.org/10.15252/emmm.201606372
Dey BK, Mueller AC, Dutta A (2014) Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 5:e944014. https://doi.org/10.4161/21541272.2014.944014
Kyrychenko S, Kyrychenko V, Badr MA et al (2015) Pivotal role of miR-448 in the development of ROS-induced cardiomyopathy. Cardiovasc Res 108:324–334. https://doi.org/10.1093/cvr/cvv238
Zhou J, Gao J, Zhang X et al (2015) microRNA-340-5p functions downstream of cardiotrophin-1 to regulate cardiac eccentric hypertrophy and heart failure via target gene dystrophin. Int Heart J 56:454–458. https://doi.org/10.1536/ihj.14-386
Quaranta MT, Spinello I, Paolillo R et al (2016) Identification of β-dystrobrevin as a direct target of miR-143: involvement in early stages of neural differentiation. PLoS One. https://doi.org/10.1371/journal.pone.0156325
Tran THT, Zhang Z, Yagi M et al (2013) Molecular characterization of an X(p21.2;q28) chromosomal inversion in a Duchenne muscular dystrophy patient with mental retardation reveals a novel long non-coding gene on Xq28. J Hum Genet 58:33–39. https://doi.org/10.1038/jhg.2012.131
Simionescu-Bankston A, Kumar A (2016) Noncoding RNAs in the regulation of skeletal muscle biology in health and disease. J Mol Med Berl Ger 94:853–866. https://doi.org/10.1007/s00109-016-1443-y
Cao H, Shao F, Li M et al (2019) Comprehensive identification of micropeptides encoded by long noncoding RNAs in human tissues. FASEB J 33:714.1. https://doi.org/10.1096/fasebj.2019.33.1_supplement.714.1
Anderson DM, Anderson KM, Chang C-L et al (2015) A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160:595–606. https://doi.org/10.1016/j.cell.2015.01.009
Makarewich CA, Munir AZ, Schiattarella GG et al (2018) The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. eLife 7:e38319. https://doi.org/10.7554/eLife.38319
Lin Y-F, Xiao M-H, Chen H-X et al (2019) A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death Dis. https://doi.org/10.1038/s41419-019-1767-y
Stein CS, Jadiya P, Zhang X et al (2018) Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep 23:3710–3720.e8. https://doi.org/10.1016/j.celrep.2018.06.002
Matsumoto A, Clohessy JG, Pandolfi PP (2017) SPAR, a lncRNA encoded mTORC1 inhibitor. Cell Cycle 16:815–816. https://doi.org/10.1080/15384101.2017.1304735
Chen B, You W, Wang Y, Shan T (2019) The regulatory role of Myomaker and Myomixer–Myomerger–Minion in muscle development and regeneration. Cell Mol Life Sci. https://doi.org/10.1007/s00018-019-03341-9
Cai B, Li Z, Ma M et al (2017) LncRNA-Six1 encodes a micropeptide to activate Six1 in Cis and is involved in cell proliferation and muscle growth. Front Physiol. https://doi.org/10.3389/fphys.2017.00230
Ma M, Cai B, Jiang L et al (2018) lncRNA-Six1 is a target of miR-1611 that functions as a ceRNA to regulate Six1 protein expression and fiber type switching in chicken myogenesis. Cells. https://doi.org/10.3390/cells7120243
Backes C, Meese E, Keller A (2016) Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol Diagn Ther 20:509–518. https://doi.org/10.1007/s40291-016-0221-4
McCarthy JJ (2008) MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta 1779:682–691. https://doi.org/10.1016/j.bbagrm.2008.03.001
Amirouche A, Jahnke VE, Lunde JA et al (2017) Muscle-specific microRNA-206 targets multiple components in dystrophic skeletal muscle representing beneficial adaptations. Am J Physiol-Cell Physiol 312:C209–C221. https://doi.org/10.1152/ajpcell.00185.2016
Liu N, Williams AH, Maxeiner JM et al (2012) microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122:2054–2065. https://doi.org/10.1172/JCI62656
Ma G, Wang Y, Li Y et al (2015) MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci 11:345–352. https://doi.org/10.7150/ijbs.10921
McCarthy JJ, Esser KA, Andrade FH (2007) MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol-Cell Physiol 293:C451–C457. https://doi.org/10.1152/ajpcell.00077.2007
Yuasa K, Hagiwara Y, Ando M et al (2008) MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct 33:163–169. https://doi.org/10.1247/csf.08022
Coenen-Stass AML, Wood MJA, Roberts TC (2017) Biomarker potential of extracellular miRNAs in duchenne muscular dystrophy. Trends Mol Med 23:989–1001. https://doi.org/10.1016/j.molmed.2017.09.002
Coenen-Stass AML, Betts CA, Lee YF et al (2016) Selective release of muscle-specific, extracellular microRNAs during myogenic differentiation. Hum Mol Genet 25:3960–3974. https://doi.org/10.1093/hmg/ddw237
Vignier N, Amor F, Fogel P et al (2013) Distinctive serum miRNA profile in mouse models of striated muscular pathologies. PLoS One 8:e55281. https://doi.org/10.1371/journal.pone.0055281
Hu J, Kong M, Ye Y et al (2014) Serum miR-206 and other muscle-specific microRNAs as non-invasive biomarkers for Duchenne muscular dystrophy. J Neurochem 129:877–883. https://doi.org/10.1111/jnc.12662
Cacchiarelli D, Legnini I, Martone J et al (2011) miRNAs as serum biomarkers for Duchenne muscular dystrophy: miRNAs as serum biomarkers for DMD. EMBO Mol Med 3:258–265. https://doi.org/10.1002/emmm.201100133
Li X, Li Y, Zhao L et al (2014) Circulating muscle-specific miRNAs in Duchenne muscular dystrophy patients. Mol Ther Nucleic Acids 3:e177. https://doi.org/10.1038/mtna.2014.29
Matsuzaka Y, Kishi S, Aoki Y et al (2014) Three novel serum biomarkers, miR-1, miR-133a, and miR-206 for Limb-girdle muscular dystrophy, Facioscapulohumeral muscular dystrophy, and Becker muscular dystrophy. Environ Health Prev Med 19:452–458. https://doi.org/10.1007/s12199-014-0405-7
Anaya-Segura M, Rangel-Villalobos H, Martínez-Cortés G et al (2016) Serum levels of MicroRNA-206 and novel mini-STR assays for carrier detection in duchenne muscular dystrophy. Int J Mol Sci 17:1334. https://doi.org/10.3390/ijms17081334
Roberts TC, Blomberg KEM, McClorey G et al (2012) Expression analysis in multiple muscle groups and serum reveals complexity in the MicroRNA transcriptome of the mdx mouse with implications for therapy. Mol Ther Nucleic Acids 1:e39. https://doi.org/10.1038/mtna.2012.26
Mizuno H, Nakamura A, Aoki Y et al (2011) Identification of muscle-specific MicroRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS One 6:e18388. https://doi.org/10.1371/journal.pone.0018388
Deng Z, Chen J-F, Wang D-Z (2011) Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskelet Disord. https://doi.org/10.1186/1471-2474-12-115
Zacharewicz E, Lamon S, Russell A (2013) MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Front Physiol 4:266. https://doi.org/10.3389/fphys.2013.00266
Gomes CPC, Oliveira GP, Madrid B et al (2014) Circulating miR-1, miR-133a, and miR-206 levels are increased after a half-marathon run. Biomark Biochem Indic Expo Response Susceptibil Chem 19:585–589. https://doi.org/10.3109/1354750X.2014.952663
Llano-Diez M, Ortez CI, Gay JA et al (2017) Digital PCR quantification of miR-30c and miR-181a as serum biomarkers for Duchenne muscular dystrophy. Neuromuscul Disord 27:15–23. https://doi.org/10.1016/j.nmd.2016.11.003
Jeanson-Leh L, Lameth J, Krimi S et al (2014) Serum profiling identifies novel muscle miRNA and cardiomyopathy-related miRNA biomarkers in golden retriever muscular dystrophy dogs and duchenne muscular dystrophy patients. Am J Pathol 184:2885–2898. https://doi.org/10.1016/j.ajpath.2014.07.021
Roberts TC, Godfrey C, McClorey G et al (2013) Extracellular microRNAs are dynamic non-vesicular biomarkers of muscle turnover. Nucleic Acids Res 41:9500–9513. https://doi.org/10.1093/nar/gkt724
Catapano F, Domingos J, Perry M et al (2018) Downregulation of miRNA-29, -23 and -21 in urine of Duchenne muscular dystrophy patients. Epigenomics. https://doi.org/10.2217/epi-2018-0022
Becker S, Florian A, Patrascu A et al (2016) Identification of cardiomyopathy associated circulating miRNA biomarkers in patients with muscular dystrophy using a complementary cardiovascular magnetic resonance and plasma profiling approach. J Cardiovasc Magn Reson. https://doi.org/10.1186/s12968-016-0244-3
Grounds MD, Terrill JR, Al-Mshhdani BA et al (2020) Biomarkers for Duchenne muscular dystrophy: myonecrosis, inflammation and oxidative stress. Dis Model Mech. https://doi.org/10.1242/dmm.043638
Cazzella V, Martone J, Pinnarò C et al (2012) Exon 45 skipping through U1-snRNA antisense molecules recovers the Dys-nNOS pathway and muscle differentiation in human DMD myoblasts. Mol Ther 20:2134–2142. https://doi.org/10.1038/mt.2012.178
Roberts TC, Johansson HJ, McClorey G et al (2015) Multi-level omics analysis in a murine model of dystrophin loss and therapeutic restoration. Hum Mol Genet 24:6756–6768. https://doi.org/10.1093/hmg/ddv381
Young CS, Hicks MR, Ermolova NV et al (2016) A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell 18:533–540. https://doi.org/10.1016/j.stem.2016.01.021
Hildyard JC, Wells DJ (2016) Investigating synthetic oligonucleotide targeting of Mir31 in Duchenne muscular dystrophy. PLoS Curr. https://doi.org/10.1371/currents.md.99d88e72634387639707601b237467d7
Heller KN, Mendell JT, Mendell JR, Rodino-Klapac LR (2017) MicroRNA-29 overexpression by adeno-associated virus suppresses fibrosis and restores muscle function in combination with micro-dystrophin. JCI Insight. https://doi.org/10.1172/jci.insight.93309
Bulaklak K, Xiao B, Qiao C et al (2018) MicroRNA-206 downregulation improves therapeutic gene expression and motor function in mdx mice. Mol Ther Nucleic Acids 12:283–293. https://doi.org/10.1016/j.omtn.2018.05.011
Perkins KJ, Davies KE (2018) Alternative utrophin mRNAs contribute to phenotypic differences between dystrophin-deficient mice and Duchenne muscular dystrophy. FEBS Lett 592:1856–1869. https://doi.org/10.1002/1873-3468.13099
Basu U, Lozynska O, Moorwood C et al (2011) Translational regulation of utrophin by miRNAs. PLoS One 6:e29376. https://doi.org/10.1371/journal.pone.0029376
Mishra MK, Loro E, Sengupta K et al (2017) Functional improvement of dystrophic muscle by repression of utrophin: let-7c interaction. PLoS One 12:e0182676. https://doi.org/10.1371/journal.pone.0182676
Aminzadeh MA, Rogers RG, Fournier M et al (2018) Exosome-mediated benefits of cell therapy in mouse and human models of duchenne muscular dystrophy. Stem Cell Rep 10:942–955. https://doi.org/10.1016/j.stemcr.2018.01.023
Roberts TC, Coenen-Stass AML, Wood MJA (2014) Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One 9:e89237. https://doi.org/10.1371/journal.pone.0089237
Acknowledgements
We gratefully thank the Associazione Centro Dino Ferrari for its support.
Funding
The project received partial support from Italian Ministry of Health to GPC.
Author information
Authors and Affiliations
Contributions
RB, FM and SC conceived and wrote the manuscript, NB and GPC revised the text.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Brusa, R., Magri, F., Bresolin, N. et al. Noncoding RNAs in Duchenne and Becker muscular dystrophies: role in pathogenesis and future prognostic and therapeutic perspectives. Cell. Mol. Life Sci. 77, 4299–4313 (2020). https://doi.org/10.1007/s00018-020-03537-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03537-4