Abstract
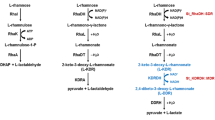
Despite physiological importance of aldonic sugar acids for living organisms, little is known about metabolic pathways of these compounds. Here, we investigated the functional diversity of homologs of L-threonic acid dehydrogenase (ThrDH; UniProt ID: Q0KBC7), an enzyme composed of two NAD-binding domains (PF14833 and PF03446). Ten ThrDH homologs with different genomic context were studied; seven new enzymatic activities were identified, such as (R)-pantoate dehydrogenase, L-altronic acid dehydrogenase, 6-deoxy-L-talonate dehydrogenase, L-idonic acid dehydrogenase, D-xylonic acid dehydrogenase, D-gluconic acid dehydrogenase, and 2-hydroxy-3-oxopantoate reductase activities. Two associated metabolic pathways were identified: L-idonic acid dehydrogenase was found to be involved in the degradation of L-idonic acid through oxidation/decarboxylation in Agrobacterium radiobacter K84, while 2-hydroxy-3-oxopantoate reductase was found to participate in D-glucarate catabolism through dehydration/cleavage in Ralstonia metallidurans CH34.








Similar content being viewed by others
Abbreviations
- EryDH:
-
D-erythronic acid dehydrogenase
- PanDH:
-
(R)-pantoate dehydrogenase
- SSN:
-
sequence similarity network
- ThrDH:
-
L-threonic acid dehydrogenase
REFERENCES
Ramachandran, S., Fontanille, P., Pandey, A., and Larroche, C. (2006) Gluconic acid: properties, applications and microbial production, Food Technol. Biotechnol., 44, 185-195.
Smirnoff, N. (2000) Ascorbic acid: metabolism and functions of a multi-facetted molecule, Curr. Opin. Plant Biol., 3, 229-235.
Vimr, E. R., Kalivoda, K. A., Deszo, E. L., and Steenbergen, S. M. (2004) Diversity of microbial sialic acid metabolism, Microbiol. Mol. Biol. R., 68, 132-153.
Wishart, D. S., Knox, C., Guo, A., Eisner, R., Young, N., et al. (2009) HMDB: a knowledgebase for the human metabolome, Nucleic Acids Res., 37, D603-D610.
Holzer, H., and Holldorf, A. (1957) Isolation of D-glycerate dehydrogenase, some properties of the enzyme and its application to the enzymic – optic determination of hydroxypyruvate in presence of pyruvate, Biochem. Z., 329, 292-312.
Dreyer, J. L. (1987) The role of iron in the activation of mannonic and altronic acid hydratases, two Fe-requiring hydrolyases, Eur. J. Biochem., 166, 623-630.
Dahms, A. S., and Donald, A. (1982) D-xylo-aldonate dehydratase, Methods Enzymol., 90, E302-E305.
Shimizu, T., Takaya, N., and Nakamura, A. (2012) An L-glucose catabolic pathway in Paracoccus species 43P, J. Biol. Chem., 287, 40448-40456.
Wichelecki, D. J., Balthazor, B. M., Chau, A. C., Vetting, M. W., Fedorov, A. A., Fedorov, E. V., Luk, T., Patskovsky, T. Y., Stead, M. B., Hillerich, B. S., Seidel, R. D., Almo, S. C., and Gerlt, J. A. (2014) Discovery of function in the enolase superfamily: d-mannonate and d-gluconate dehydratases in the d-mannonate dehydratase subgroup, Biochemistry, 53, 2722-2731.
Zhang, X., Carter, M. S., Vetting, M. W., Francisco, B. S., Zhao, S., Al-Obaidi, N. F., Solbiatia, J. O., Thiavilled, J. J., Crecy-Lagard, V., Jacobson, M. P., Almo, S. C., and Gerlt, J. A. (2016) Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars, Proc. Natl. Acad. Sci. USA, 113, E4161-E4169.
Gerlt, J. A., Bouvier, J. T., Davidson, D. B., Imker, H. J., Sadkhin, B., Slater, D. R., and Whalen, K. L. (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks, Biochim. Biophys. Acta, 1854, 1019-1037.
Luo, S., and Huang, H. (2018) Discovering a new catabolic pathway of D-ribonate in Mycobacterium smegmatis, Biochem. Biophys. Res. Commun., 505, 1107-1111.
Aghaie, A., Lechaplais, C., Sirven, P., Tricot, S., Besnard-Gonnet, M., Muselet, D., Berardinis, V., Kreimeyer, A., Gyapay, G., Salanoubat, M., and Perret, A. (2008) New insights into the alternative D-glucarate degradation pathway, J. Biol. Chem., 283, 15638-15646.
Severi, E., Hood, D. W., and Thomas, G. H. (2007) Sialic acid utilization by bacterial pathogens, Microbiology, 153, 2817-2822.
Adachi, O., Hours, R. A., Shinagawa, E., Akakabe, Y., Ykushi, T., and Matsushita, K. (2011) Enzymatic synthesis of 4-pentulosonate (4-keto-D-pentonate) from D-aldopentose and D-pentonate by two different pathways using membrane enzymes of acetic acid bacteria, Biosci. Biotechnol. Biochem., 75, 2418-2420.
Arrigoni, O., and De Tulio, M. C. (2002) Ascorbic acid: much more than just an antioxidant, Biochim. Biophys. Acta, 1569, 1-9.
Yew, W. S., Fedorov, A. A., Fedorov, E. V., Rakus, J. F., Pierce, R. W., Almo, S. C., and Gerlt, J. A. (2006) Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris, Biochemistry, 45, 14582-14597.
Tchigvintsev, A., Singer, A., Brown, G., Flick, R., Evdokimova, E., Tan, K., Gonzalez, C. F., Savchenko, A., and Yakunin, A. F. (2012) Biochemical and structural studies of uncharacterized protein PA0743 from Pseudomonas aeruginosa revealed NAD-dependent L-serine dehydrogenase, J. Biol. Chem., 287, 1874-1883.
Srikalaivani, R., Singh, A., Vijayan, M., and Surolia, A. (2018) Structure, interactions and action of Mycobacterium tuberculosis 3-hydroxyisobutyric acid dehydrogenase, Biochem. J., 475, 2457-2471.
Zheng, R., and Blanchard, J. S. (2000) Kinetic and mechanistic analysis of the E. coli panE-encoded ketopantoate reductase, Biochemistry, 39, 3708-3717.
Webb, M. E., Smith, A. G., and Abell, C. (2004) Biosynthesis of pantothenate, Nat. Prod. Rep., 21, 695-721.
Grostern, A., Sales, C. M., Zhuang, W. Q., Erbilgin, O., and Alvarez-Cohen, L. (2012) Glyoxylate metabolism is a key feature of the metabolic degradation of 1,4-dioxane by Pseudonocardia dioxanivorans strain CB1190, Appl. Environ. Microbiol., 78, 3298-3308.
Funding
This work was supported by the National Natural Science Foundation of China (project No. 31970087) and the start-up grant from the South China Normal University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This article does not contain description of studies with human participants or animals performed by any of the authors. The authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, C., Liu, Y., Wu, X. et al. Substrate Diversity of L-Threonic Acid Dehydrogenase Homologs. Biochemistry Moscow 85, 463–471 (2020). https://doi.org/10.1134/S0006297920040069
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920040069