Abstract
Background
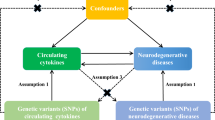
Multiple sclerosis (MS) is a complex neurological disease and chronic inflammatory disease. Until now, observational studies have reported positive association between serum interleukin-6 (IL-6) levels and MS risk. In order to develop effective therapies, we should establish the causal link between IL-6 signaling and MS. However, it is currently unknown whether IL-6 signaling is causally associated with the risk of MS.
Methods
Here, we selected the increased soluble IL-6R (s-IL-6R) levels as the indirect markers for reduced IL-6 signaling, and performed a Mendelian randomization (MR) study using the rs2228145 variant as the instrumental variable to evaluate and quantify the effect of IL-6 signaling on the risk of MS. To be a comparison, we also evaluated the causal association of IL-6 signaling with the risk of other three neurodegenerative diseases including Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS).
Results
We found that the increased s-IL-6R levels (per 1 standard deviation) were significantly associated with decreased MS risk (OR = 0.96, 95% CI 0.94–0.98, P = 1.69E-04), but not associated with the risk of AD (OR = 1.01, 95% CI 0.92–1.11, P = 0.835), PD (OR = 0.94, 95% CI 0.84–1.05, P = 0.261), or ALS (OR = 1.00, 95% CI 0.92–1.10, P = 0.9411).
Conclusion
Our findings have the similar directional effects to an existing humanized anti-IL-6R monoclonal antibody Tocilizumab which could bind to the IL-6 binding site of human IL-6R and competitively inhibit IL-6 signaling. Hence, we provided genetic evidence that inhibiting the IL-6 signaling such as tocilizumab treatment might represent a novel therapy for MS.
Similar content being viewed by others
Data availability
All relevant data are within the paper. The authors confirm that all data underlying the findings are either fully available without restriction through consortia websites or may be made available from consortia upon request. Social Science Genetic Association Consortium (SSGAC): https://www.thessgac.org/data; type 2 diabetes from DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium: http://diagram-consortium.org/downloads.html; Tobacco and Genetics Consortium (TGC): http://www.med.unc.edu/pgc/downloads; Ukbiobank: http://www.ukbiobank.ac.uk/scientists-3/genetic-data/;https://sites.google.com/broadinstitute.org/ukbbgwasresults/home?authuser=0;http://geneatlas.roslin.ed.ac.uk; International MS Genetics Consortium (IMSGC): http://imsgc.net/publications/.
References
Liu G, Zhang F, Jiang Y, Hu Y, Gong Z, Liu S, Chen X, Jiang Q, Hao J (2017) Integrating genome-wide association studies and gene expression data highlights dysregulated multiple sclerosis risk pathways. Mult Scler 23(2):205–212
Liu G, Zhang F, Hu Y, Jiang Y, Gong Z, Liu S, Chen X, Jiang Q, Hao J (2017) Genetic variants and multiple sclerosis risk gene SLC9A9 expression in distinct human brain regions. Mol Neurobiol 54(9):6820–6826
Liu G, Hu Y, Jin S, Jiang Q (2017) Genetic variant rs763361 regulates multiple sclerosis CD226 gene expression. Proc Natl Acad Sci U S A 114(6):E906–E907
Zhang Y, Wang L, Jia H, Liao M, Chen X, Xu J, Bao Y, Liu G (2018) Genetic variants regulate NR1H3 expression and contribute to multiple sclerosis risk. J Neurol Sci 390:162–165
Liu G, Jin S, Jiang Q (2019) Interleukin-6 receptor and inflammatory bowel disease: a Mendelian randomization study. Gastroenterology 156(3):823–824
Parisinos CA, Serghiou S, Katsoulis M, George MJ, Patel RS, Hemingway H, Hingorani AD (2018) Variation in interleukin 6 receptor gene associates with risk of Crohn’s disease and ulcerative colitis. Gastroenterology 155(2):303–306 e302
Lambertsen KL, Finsen B, Clausen BH (2019) Post-stroke inflammation-target or tool for therapy? Acta Neuropathol 137(5):693–714
Kang S, Tanaka T, Narazaki M, Kishimoto T (2019) Targeting interleukin-6 signaling in clinic. Immunity 50(4):1007–1023
Swerdlow DI, Holmes MV, Kuchenbaecker KB, Engmann JE, Shah T, Sofat R, Guo Y, Chung C, Peasey A, Pfister R, Mooijaart SP, Ireland HA, Leusink M, Langenberg C, Li KW, Palmen J, Howard P, Cooper JA, Drenos F, Hardy J, Nalls MA, Li YR, Lowe G, Stewart M, Bielinski SJ, Peto J, Timpson NJ, Gallacher J, Dunlop M, Houlston R, Tomlinson I, Tzoulaki I, Luan J, Boer JM, Forouhi NG, Onland-Moret NC, van der Schouw YT, Schnabel RB, Hubacek JA, Kubinova R, Baceviciene M, Tamosiunas A, Pajak A, Topor-Madry R, Malyutina S, Baldassarre D, Sennblad B, Tremoli E, de Faire U, Ferrucci L, Bandenelli S, Tanaka T, Meschia JF, Singleton A, Navis G, Mateo Leach I, Bakker SJ, Gansevoort RT, Ford I, Epstein SE, Burnett MS, Devaney JM, Jukema JW, Westendorp RG, Jan de Borst G, van der Graaf Y, de Jong PA, Mailand-van der Zee AH, Klungel OH, de Boer A, Doevendans PA, Stephens JW, Eaton CB, Robinson JG, Manson JE, Fowkes FG, Frayling TM, Price JF, Whincup PH, Morris RW, Lawlor DA, Smith GD, Ben-Shlomo Y, Redline S, Lange LA, Kumari M, Wareham NJ, Verschuren WM, Benjamin EJ, Whittaker JC, Hamsten A, Dudbridge F, Delaney JA, Wong A, Kuh D, Hardy R, Castillo BA, Connolly JJ, van der Harst P, Brunner EJ, Marmot MG, Wassel CL, Humphries SE, Talmud PJ, Kivimaki M, Asselbergs FW, Voevoda M, Bobak M, Pikhart H, Wilson JG, Hakonarson H, Reiner AP, Keating BJ, Sattar N, Hingorani AD, Casas JP (2012) The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet 379(9822):1214–1224
Stelmasiak Z, Koziol-Montewka M, Dobosz B, Rejdak K, Bartosik-Psujek H, Mitosek-Szewczyk K, Belniak-Legiec E (2000) Interleukin-6 concentration in serum and cerebrospinal fluid in multiple sclerosis patients. Med Sci Monit 6(6):1104–1108
Chen YC, Yang X, Miao L, Liu ZG, Li W, Zhao ZX, Sun XJ, Jiang GX, Chen SD, Cheng Q (2012) Serum level of interleukin-6 in Chinese patients with multiple sclerosis. J Neuroimmunol 249(1–2):109–111
Kallaur AP, Oliveira SR, Colado Simao AN, Delicato de Almeida ER, Kaminami Morimoto H, Lopes J, de Carvalho Jennings Pereira WL, Marques Andrade R, Muliterno Pelegrino L, Donizete Borelli S, Kaimen-Maciel DR, Reiche EM (2013) Cytokine profile in relapsing-remitting multiple sclerosis patients and the association between progression and activity of the disease. Mol Med Rep 7(3):1010–1020
Liu G, Zhao Y, Jin S, Hu Y, Wang T, Tian R, Han Z, Xu D, Jiang Q (2018) Circulating vitamin E levels and Alzheimer’s disease: a Mendelian randomization study. Neurobiol Aging 72:189 e181–189 e189
Zhao JV, Schooling CM (2019) Effect of linoleic acid on ischemic heart disease and its risk factors: a Mendelian randomization study. BMC Med 17(1):61
Zhao JV, Kwok MK, Schooling CM (2019) Effect of glutamate and aspartate on ischemic heart disease, blood pressure, and diabetes: a Mendelian randomization study. Am J Clin Nutr 109(4):1197–1206
Luo S, Au Yeung SL, Zhao JV, Burgess S, Schooling CM (2019) Association of genetically predicted testosterone with thromboembolism, heart failure, and myocardial infarction: mendelian randomisation study in UK Biobank. BMJ 364:l476
Emdin CA, Khera AV, Kathiresan S (2017) Mendelian Randomization. JAMA 318(19):1925–1926
Larsson SC, Traylor M, Burgess S, Boncoraglio GB, Jern C, Michaelsson K, Markus HS (2019) Serum magnesium and calcium levels in relation to ischemic stroke: Mendelian randomization study. Neurology 92(9):e944–e950
Grover S, Lill CM, Kasten M, Klein C, Del Greco MF, Konig IR (2019) Risky behaviors and Parkinson disease: a mendelian randomization study. Neurology 93(15):e1412–e1424
Wang L, Qiao Y, Zhang H, Zhang Y, Hua J, Jin S, Liu G (2020) Circulating vitamin D levels and Alzheimer’s disease: a Mendelian randomization study in the IGAP and UK Biobank. J Alzheimers Dis 73(2):609–618
Zhang H, Wang T, Han Z, Wang L, Zhang Y, Liu G (2020) Impact of vitamin D binding protein levels on Alzheimer’s disease: a Mendelian randomization study. J Alzheimers Dis 74(3):991–998
Sarwar N, Butterworth AS, Freitag DF, Gregson J, Willeit P, Gorman DN, Gao P, Saleheen D, Rendon A, Nelson CP, Braund PS, Hall AS, Chasman DI, Tybjaerg-Hansen A, Chambers JC, Benjamin EJ, Franks PW, Clarke R, Wilde AA, Trip MD, Steri M, Witteman JC, Qi L, van der Schoot CE, de Faire U, Erdmann J, Stringham HM, Koenig W, Rader DJ, Melzer D, Reich D, Psaty BM, Kleber ME, Panagiotakos DB, Willeit J, Wennberg P, Woodward M, Adamovic S, Rimm EB, Meade TW, Gillum RF, Shaffer JA, Hofman A, Onat A, Sundstrom J, Wassertheil-Smoller S, Mellstrom D, Gallacher J, Cushman M, Tracy RP, Kauhanen J, Karlsson M, Salonen JT, Wilhelmsen L, Amouyel P, Cantin B, Best LG, Ben-Shlomo Y, Manson JE, Davey-Smith G, de Bakker PI, O'Donnell CJ, Wilson JF, Wilson AG, Assimes TL, Jansson JO, Ohlsson C, Tivesten A, Ljunggren O, Reilly MP, Hamsten A, Ingelsson E, Cambien F, Hung J, Thomas GN, Boehnke M, Schunkert H, Asselbergs FW, Kastelein JJ, Gudnason V, Salomaa V, Harris TB, Kooner JS, Allin KH, Nordestgaard BG, Hopewell JC, Goodall AH, Ridker PM, Holm H, Watkins H, Ouwehand WH, Samani NJ, Kaptoge S, Di Angelantonio E, Harari O, Danesh J (2012) Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet 379(9822):1205–1213
Emdin CA, Khera AV, Natarajan P, Klarin D, Zekavat SM, Hsiao AJ, Kathiresan S (2017) Genetic association of waist-to-hip ratio with cardiometabolic traits, type 2 diabetes, and coronary heart disease. JAMA 317(6):626–634
International Multiple Sclerosis Genetics Consortium (2018) Low-frequency and rare-coding variation contributes to multiple sclerosis risk. Cell 175(6):1679–1687.e7
Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, Frizatti A, Chouraki V, Martin ER, Sleegers K, Badarinarayan N, Jakobsdottir J, Hamilton-Nelson KL, Moreno-Grau S, Olaso R, Raybould R, Chen Y, Kuzma AB, Hiltunen M, Morgan T, Ahmad S, Vardarajan BN, Epelbaum J, Hoffmann P, Boada M, Beecham GW, Garnier JG, Harold D, Fitzpatrick AL, Valladares O, Moutet ML, Gerrish A, Smith AV, Qu L, Bacq D, Denning N, Jian X, Zhao Y, Del Zompo M, Fox NC, Choi SH, Mateo I, Hughes JT, Adams HH, Malamon J, Sanchez-Garcia F, Patel Y, Brody JA, Dombroski BA, Naranjo MCD, Daniilidou M, Eiriksdottir G, Mukherjee S, Wallon D, Uphill J, Aspelund T, Cantwell LB, Garzia F, Galimberti D, Hofer E, Butkiewicz M, Fin B, Scarpini E, Sarnowski C, Bush WS, Meslage S, Kornhuber J, White CC, Song Y, Barber RC, Engelborghs S, Sordon S, Voijnovic D, Adams PM, Vandenberghe R, Mayhaus M, Cupples LA, Albert MS, De Deyn PP, Gu W, Himali JJ, Beekly D, Squassina A, Hartmann AM, Orellana A, Blacker D, Rodriguez-Rodriguez E, Lovestone S, Garcia ME, Doody RS, Munoz-Fernadez C, Sussams R, Lin H, Fairchild TJ, Benito YA, Holmes C, Karamujic-Comic H, Frosch MP, Thonberg H, Maier W, Roschupkin G, Ghetti B, Giedraitis V, Kawalia A, Li S, Huebinger RM, Kilander L, Moebus S, Hernandez I, Kamboh MI, Brundin R, Turton J, Yang Q, Katz MJ, Concari L, Lord J, Beiser AS, Keene CD, Helisalmi S, Kloszewska I, Kukull WA, Koivisto AM, Lynch A, Tarraga L, Larson EB, Haapasalo A, Lawlor B, Mosley TH, Lipton RB, Solfrizzi V, Gill M, Longstreth WT Jr, Montine TJ, Frisardi V, Diez-Fairen M, Rivadeneira F, Petersen RC, Deramecourt V, Alvarez I, Salani F, Ciaramella A, Boerwinkle E, Reiman EM, Fievet N, Rotter JI, Reisch JS, Hanon O, Cupidi C, Andre Uitterlinden AG, Royall DR, Dufouil C, Maletta RG, de Rojas I, Sano M, Brice A, Cecchetti R, George-Hyslop PS, Ritchie K, Tsolaki M, Tsuang DW, Dubois B, Craig D, Wu CK, Soininen H, Avramidou D, Albin RL, Fratiglioni L, Germanou A, Apostolova LG, Keller L, Koutroumani M, Arnold SE, Panza F, Gkatzima O, Asthana S, Hannequin D, Whitehead P, Atwood CS, Caffarra P, Hampel H, Quintela I, Carracedo A, Lannfelt L, Rubinsztein DC, Barnes LL, Pasquier F, Frolich L, Barral S, McGuinness B, Beach TG, Johnston JA, Becker JT, Passmore P, Bigio EH, Schott JM, Bird TD, Warren JD, Boeve BF, Lupton MK, Bowen JD, Proitsi P, Boxer A, Powell JF, Burke JR, Kauwe JSK, Burns JM, Mancuso M, Buxbaum JD, Bonuccelli U, Cairns NJ, McQuillin A, Cao C, Livingston G, Carlson CS, Bass NJ, Carlsson CM, Hardy J, Carney RM, Bras J, Carrasquillo MM, Guerreiro R, Allen M, Chui HC, Fisher E, Masullo C, Crocco EA, DeCarli C, Bisceglio G, Dick M, Ma L, Duara R, Graff-Radford NR, Evans DA, Hodges A, Faber KM, Scherer M, Fallon KB, Riemenschneider M, Fardo DW, Heun R, Farlow MR, Kolsch H, Ferris S, Leber M, Foroud TM, Heuser I, Galasko DR, Giegling I, Gearing M, Hull M, Geschwind DH, Gilbert JR, Morris J, Green RC, Mayo K, Growdon JH, Feulner T, Hamilton RL, Harrell LE, Drichel D, Honig LS, Cushion TD, Huentelman MJ, Hollingworth P, Hulette CM, Hyman BT, Marshall R, Jarvik GP, Meggy A, Abner E, Menzies GE, Jin LW, Leonenko G, Real LM, Jun GR, Baldwin CT, Grozeva D, Karydas A, Russo G, Kaye JA, Kim R, Jessen F, Kowall NW, Vellas B, Kramer JH, Vardy E, LaFerla FM, Jockel KH, Lah JJ, Dichgans M, Leverenz JB, Mann D, Levey AI, Pickering-Brown S, Lieberman AP, Klopp N, Lunetta KL, Wichmann HE, Lyketsos CG, Morgan K, Marson DC, Brown K, Martiniuk F, Medway C, Mash DC, Nothen MM, Masliah E, Hooper NM, McCormick WC, Daniele A, McCurry SM, Bayer A, McDavid AN, Gallacher J, McKee AC, van den Bussche H, Mesulam M, Brayne C, Miller BL, Riedel-Heller S, Miller CA, Miller JW, Al-Chalabi A, Morris JC, Shaw CE, Myers AJ, Wiltfang J, O'Bryant S, Olichney JM, Alvarez V, Parisi JE, Singleton AB, Paulson HL, Collinge J, Perry WR, Mead S, Peskind E, Cribbs DH, Rossor M, Pierce A, Ryan NS, Poon WW, Nacmias B, Potter H, Sorbi S, Quinn JF, Sacchinelli E, Raj A, Spalletta G, Raskind M, Caltagirone C, Bossu P, Orfei MD, Reisberg B, Clarke R, Reitz C, Smith AD, Ringman JM, Warden D, Roberson ED, Wilcock G, Rogaeva E, Bruni AC, Rosen HJ, Gallo M, Rosenberg RN, Ben-Shlomo Y, Sager MA, Mecocci P, Saykin AJ, Pastor P, Cuccaro ML, Vance JM, Schneider JA, Schneider LS, Slifer S, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tang M, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Saba Y, Pilotto A, Bullido MJ, Peters O, Crane PK, Bennett D, Bosco P, Coto E, Boccardi V, De Jager PL, Lleo A, Warner N, Lopez OL, Ingelsson M, Deloukas P, Cruchaga C, Graff C, Gwilliam R, Fornage M, Goate AM, Sanchez-Juan P, Kehoe PG, Amin N, Ertekin-Taner N, Berr C, Debette S, Love S, Launer LJ, Younkin SG, Dartigues JF, Corcoran C, Ikram MA, Dickson DW, Nicolas G, Campion D, Tschanz J, Schmidt H, Hakonarson H, Clarimon J, Munger R, Schmidt R, Farrer LA, Van Broeckhoven C, COD M, AL DS, Jones L, Haines JL, Deleuze JF, Owen MJ, Gudnason V, Mayeux R, Escott-Price V, Psaty BM, Ramirez A, Wang LS, Ruiz A, van Duijn CM, Holmans PA, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Lambert JC, Pericak-Vance MA (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 51(3):414–430
Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, Bras J, Young E, Coelln RV, Simón-Sánchez J, Schulte C, Sharma M, Krohn L, Pihlstrom L, Siitonen A, Hirotaka I (2019) Expanding Parkinson’s disease genetics: novel risk loci, genomic context, causal insights and heritable risk. bioRxiv. https://doi.org/10.1101/388165
Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, Dominov JA, Kenna BJ, Nalls MA, Keagle P, Rivera AM, van Rheenen W, Murphy NA, van Vugt J, Geiger JT, Van der Spek RA, Pliner HA, Shankaracharya SBN, Marangi G, Topp SD, Abramzon Y, Gkazi AS, Eicher JD, Kenna A, Mora G, Calvo A, Mazzini L, Riva N, Mandrioli J, Caponnetto C, Battistini S, Volanti P, La Bella V, Conforti FL, Borghero G, Messina S, Simone IL, Trojsi F, Salvi F, Logullo FO, D'Alfonso S, Corrado L, Capasso M, Ferrucci L, Moreno CAM, Kamalakaran S, Goldstein DB, Gitler AD, Harris T, Myers RM, Phatnani H, Musunuri RL, Evani US, Abhyankar A, Zody MC, Kaye J, Finkbeiner S, Wyman SK, LeNail A, Lima L, Fraenkel E, Svendsen CN, Thompson LM, Van Eyk JE, Berry JD, Miller TM, Kolb SJ, Cudkowicz M, Baxi E, Benatar M, Taylor JP, Rampersaud E, Wu G, Wuu J, Lauria G, Verde F, Fogh I, Tiloca C, Comi GP, Soraru G, Cereda C, Corcia P, Laaksovirta H, Myllykangas L, Jansson L, Valori M, Ealing J, Hamdalla H, Rollinson S, Pickering-Brown S, Orrell RW, Sidle KC, Malaspina A, Hardy J, Singleton AB, Johnson JO, Arepalli S, Sapp PC, McKenna-Yasek D, Polak M, Asress S, Al-Sarraj S, King A, Troakes C, Vance C, de Belleroche J, Baas F, Ten Asbroek A, Munoz-Blanco JL, Hernandez DG, Ding J, Gibbs JR, Scholz SW, Floeter MK, Campbell RH, Landi F, Bowser R, Pulst SM, Ravits JM, MacGowan DJL, Kirby J, Pioro EP, Pamphlett R, Broach J, Gerhard G, Dunckley TL, Brady CB, Kowall NW, Troncoso JC, Le Ber I, Mouzat K, Lumbroso S, Heiman-Patterson TD, Kamel F, Van Den Bosch L, Baloh RH, Strom TM, Meitinger T, Shatunov A, Van Eijk KR, de Carvalho M, Kooyman M, Middelkoop B, Moisse M, McLaughlin RL, Van Es MA, Weber M, Boylan KB, Van Blitterswijk M, Rademakers R, Morrison KE, Basak AN, Mora JS, Drory VE, Shaw PJ, Turner MR, Talbot K, Hardiman O, Williams KL, Fifita JA, Nicholson GA, Blair IP, Rouleau GA, Esteban-Perez J, Garcia-Redondo A, Al-Chalabi A, Rogaeva E, Zinman L, Ostrow LW, Maragakis NJ, Rothstein JD, Simmons Z, Cooper-Knock J, Brice A, Goutman SA, Feldman EL, Gibson SB, Taroni F, Ratti A, Gellera C, Van Damme P, Robberecht W, Fratta P, Sabatelli M, Lunetta C, Ludolph AC, Andersen PM, Weishaupt JH, Camu W, Trojanowski JQ, Van Deerlin VM, Brown RH Jr, van den Berg LH, Veldink JH, Harms MB, Glass JD, Stone DJ, Tienari P, Silani V, Chio A, Shaw CE, Traynor BJ, Landers JE (2018) Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97(6):1268–1283 e1266
Dziedzic T, Slowik A, Szczudlik A (2003) Interleukin-6 and stroke: cerebral ischemia versus nonspecific factors influencing interleukin-6. Stroke 34(12):e229–e230 author reply e229-230
Belbasis L, Bellou V, Evangelou E, Ioannidis JP, Tzoulaki I (2015) Environmental risk factors and multiple sclerosis: an umbrella review of systematic reviews and meta-analyses. Lancet Neurol 14(3):263–273
Tobacco and Genetics Consortium (2010) Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42(5):441–447
Scott RA, Scott LJ, Magi R, Marullo L, Gaulton KJ, Kaakinen M, Pervjakova N, Pers TH, Johnson AD, Eicher JD, Jackson AU, Ferreira T, Lee Y, Ma C, Steinthorsdottir V, Thorleifsson G, Qi L, Van Zuydam NR, Mahajan A, Chen H, Almgren P, Voight BF, Grallert H, Muller-Nurasyid M, Ried JS, Rayner NW, Robertson N, Karssen LC, van Leeuwen EM, Willems SM, Fuchsberger C, Kwan P, Teslovich TM, Chanda P, Li M, Lu Y, Dina C, Thuillier D, Yengo L, Jiang L, Sparso T, Kestler HA, Chheda H, Eisele L, Gustafsson S, Franberg M, Strawbridge RJ, Benediktsson R, Hreidarsson AB, Kong A, Sigurethsson G, Kerrison ND, Luan J, Liang L, Meitinger T, Roden M, Thorand B, Esko T, Mihailov E, Fox C, Liu CT, Rybin D, Isomaa B, Lyssenko V, Tuomi T, Couper DJ, Pankow JS, Grarup N, Have CT, Jorgensen ME, Jorgensen T, Linneberg A, Cornelis MC, van Dam RM, Hunter DJ, Kraft P, Sun Q, Edkins S, Owen KR, Perry JRB, Wood AR, Zeggini E, Tajes-Fernandes J, Abecasis GR, Bonnycastle LL, Chines PS, Stringham HM, Koistinen HA, Kinnunen L, Sennblad B, Muhleisen TW, Nothen MM, Pechlivanis S, Baldassarre D, Gertow K, Humphries SE, Tremoli E, Klopp N, Meyer J, Steinbach G, Wennauer R, Eriksson JG, Mnnisto S, Peltonen L, Tikkanen E, Charpentier G, Eury E, Lobbens S, Gigante B, Leander K, McLeod O, Bottinger EP, Gottesman O, Ruderfer D, Bluher M, Kovacs P, Tonjes A, Maruthur NM, Scapoli C, Erbel R, Jockel KH, Moebus S, de Faire U, Hamsten A, Stumvoll M, Deloukas P, Donnelly PJ, Frayling TM, Hattersley AT, Ripatti S, Salomaa V, Pedersen NL, Boehm BO, Bergman RN, Collins FS, Mohlke KL, Tuomilehto J, Hansen T, Pedersen O, Barroso I, Lannfelt L, Ingelsson E, Lind L, Lindgren CM, Cauchi S, Froguel P, Loos RJF, Balkau B, Boeing H, Franks PW, Barricarte Gurrea A, Palli D, van der Schouw YT, Altshuler D, Groop LC, Langenberg C, Wareham NJ, Sijbrands E, van Duijn CM, Florez JC, Meigs JB, Boerwinkle E, Gieger C, Strauch K, Metspalu A, Morris AD, Palmer CNA, Hu FB, Thorsteinsdottir U, Stefansson K, Dupuis J, Morris AP, Boehnke M, McCarthy MI, Prokopenko I (2017) An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 66(11):2888–2902
Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, Meddens SF, Linner RK, Rietveld CA, Derringer J, Gratten J, Lee JJ, Liu JZ, de Vlaming R, Ahluwalia TS, Buchwald J, Cavadino A, Frazier-Wood AC, Furlotte NA, Garfield V, Geisel MH, Gonzalez JR, Haitjema S, Karlsson R, van der Laan SW, Ladwig KH, Lahti J, van der Lee SJ, Lind PA, Liu T, Matteson L, Mihailov E, Miller MB, Minica CC, Nolte IM, Mook-Kanamori D, van der Most PJ, Oldmeadow C, Qian Y, Raitakari O, Rawal R, Realo A, Rueedi R, Schmidt B, Smith AV, Stergiakouli E, Tanaka T, Taylor K, Thorleifsson G, Wedenoja J, Wellmann J, Westra HJ, Willems SM, Zhao W, Amin N, Bakshi A, Bergmann S, Bjornsdottir G, Boyle PA, Cherney S, Cox SR, Davies G, Davis OS, Ding J, Direk N, Eibich P, Emeny RT, Fatemifar G, Faul JD, Ferrucci L, Forstner AJ, Gieger C, Gupta R, Harris TB, Harris JM, Holliday EG, Hottenga JJ, De Jager PL, Kaakinen MA, Kajantie E, Karhunen V, Kolcic I, Kumari M, Launer LJ, Franke L, Li-Gao R, Liewald DC, Koini M, Loukola A, Marques-Vidal P, Montgomery GW, Mosing MA, Paternoster L, Pattie A, Petrovic KE, Pulkki-Raback L, Quaye L, Raikkonen K, Rudan I, Scott RJ, Smith JA, Sutin AR, Trzaskowski M, Vinkhuyzen AE, Yu L, Zabaneh D, Attia JR, Bennett DA, Berger K, Bertram L, Boomsma DI, Snieder H, Chang SC, Cucca F, Deary IJ, van Duijn CM, Eriksson JG, Bultmann U, de Geus EJ, Groenen PJ, Gudnason V, Hansen T, Hartman CA, Haworth CM, Hayward C, Heath AC, Hinds DA, Hypponen E, Iacono WG, Jarvelin MR, Jockel KH, Kaprio J, Kardia SL, Keltikangas-Jarvinen L, Kraft P, Kubzansky LD, Lehtimaki T, Magnusson PK, Martin NG, McGue M, Metspalu A, Mills M, de Mutsert R, Oldehinkel AJ, Pasterkamp G, Pedersen NL, Plomin R, Polasek O, Power C, Rich SS, Rosendaal FR, den Ruijter HM, Schlessinger D, Schmidt H, Svento R, Schmidt R, Alizadeh BZ, Sorensen TI, Spector TD, Starr JM, Stefansson K, Steptoe A, Terracciano A, Thorsteinsdottir U, Thurik AR, Timpson NJ, Tiemeier H, Uitterlinden AG, Vollenweider P, Wagner GG, Weir DR, Yang J, Conley DC, Smith GD, Hofman A, Johannesson M, Laibson DI, Medland SE, Meyer MN, Pickrell JK, Esko T, Krueger RF, Beauchamp JP, Koellinger PD, Benjamin DJ, Bartels M, Cesarini D (2016) Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 48(6):624–633
Canela-Xandri O, Rawlik K, Tenesa A (2018) An atlas of genetic associations in UK Biobank. Nat Genet 50(11):1593–1599
Ireland SJ, Monson NL, Davis LS (2015) Seeking balance: potentiation and inhibition of multiple sclerosis autoimmune responses by IL-6 and IL-10. Cytokine 73(2):236–244
Padberg F, Feneberg W, Schmidt S, Schwarz MJ, Korschenhausen D, Greenberg BD, Nolde T, Muller N, Trapmann H, Konig N, Moller HJ, Hampel H (1999) CSF and serum levels of soluble interleukin-6 receptors (sIL-6R and sgp130), but not of interleukin-6 are altered in multiple sclerosis. J Neuroimmunol 99(2):218–223
Stelmasiak Z, Koziol-Montewka M, Dobosz B, Rejdak K (2001) IL-6 and sIL-6R concentration in the cerebrospinal fluid and serum of MS patients. Med Sci Monit 7(5):914–918
Schneider A, Long SA, Cerosaletti K, Ni CT, Samuels P, Kita M, Buckner JH (2013) In active relapsing-remitting multiple sclerosis, effector T cell resistance to adaptive T(regs) involves IL-6-mediated signaling. Sci Transl Med 5(170):170ra115
Khaibullin T, Ivanova V, Martynova E, Cherepnev G, Khabirov F, Granatov E, Rizvanov A, Khaiboullina S (2017) Elevated levels of proinflammatory cytokines in cerebrospinal fluid of multiple sclerosis patients. Front Immunol 8:531
Rosa M, Chignon A, Li Z, Boulanger M-C, Arsenault BJ, Bossé Y, Thériault S, Mathieu P (2019) A Mendelian randomization study of IL6 signaling in cardiovascular diseases, immune-related disorders and longevity. npj Genomic Med:4–23
Nishimoto N, Terao K, Mima T, Nakahara H, Takagi N, Kakehi T (2008) Mechanisms and pathologic significances in increase in serum interleukin-6 (IL-6) and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood 112(10):3959–3964
Arakia M, Nakamura M, Sato W, Takahashi Y, Yamamura T (2017) Potential benefits of the anti-IL-6 receptor antibody tocilizumab in multiple sclerosis patients with high plasmablast frequency. J Neurol Sci 381(Supplement):131
Sato H, Kobayashi D, Abe A, Ito S, Ishikawa H, Nakazono K, Murasawa A, Kuroda T, Nakano M, Narita I (2014) Tocilizumab treatment safety in rheumatoid arthritis in a patient with multiple sclerosis: a case report. BMC Res Notes 7:641
Schett G (2018) Physiological effects of modulating the interleukin-6 axis. Rheumatology (Oxford) 57(suppl_2):ii43–ii50
Rothaug M, Becker-Pauly C, Rose-John S (2016) The role of interleukin-6 signaling in nervous tissue. Biochim Biophys Acta 1863(6 Pt A):1218–1227
Gobel K, Ruck T, Meuth SG (2018) Cytokine signaling in multiple sclerosis: lost in translation. Mult Scler 24(4):432–439
Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS (2018) Genomic atlas of the human plasma proteome. Nature 558(7708):73–79
Larsson SC, Burgess S, Michaelsson K (2017) Association of genetic variants related to serum calcium levels with coronary artery disease and myocardial infarction. JAMA 318(4):371–380
Larsson SC, Traylor M, Malik R, Dichgans M, Burgess S, Markus HS (2017) Modifiable pathways in Alzheimer’s disease: Mendelian randomisation analysis. BMJ 359:j5375
Acknowledgments
We thank International Multiple Sclerosis Genetics Consortium, International Genomics of Alzheimer’s Project (IGAP), International Parkinson’s Disease Genomics Consortium, Tobacco and Genetics Consortium, and the SUNLIGHT consortium for the GWAS summary statistics. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. We thank Longcai Wang and Yan Zhang for their contribution to this revision.
Funding
The i-Select chips was funded by the French National Foundation on AD and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2, and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant no. 503480), Alzheimer’s Research UK (Grant no. 503176), the Wellcome Trust (Grant no. 082604/2/07/Z), and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant no. 01GI0102, 01GI0711, and 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01-AG-12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC-10-196728.
Author information
Authors and Affiliations
Contributions
GYL and HHZ conceived and initiated the project. GYL and HHZ analyzed the data and wrote the first draft of the manuscript. All authors contributed to the interpretation of the results and critical revision of the manuscript for important intellectual content and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This article contains human participants collected by several studies performed by previous studies. All participants gave informed consent in all the corresponding original studies, as described in the “Materials and methods” section. Here, our study is based on the publicly available, large-scale GWAS summary datasets, and not the individual-level data. Hence, ethical approval was not sought.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, H., Wang, T., Han, Z. et al. Mendelian randomization study to evaluate the effects of interleukin-6 signaling on four neurodegenerative diseases. Neurol Sci 41, 2875–2882 (2020). https://doi.org/10.1007/s10072-020-04381-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-020-04381-x