Abstract
Purpose
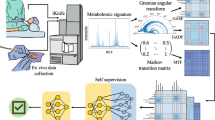
Basal cell carcinoma (BCC) is the most commonly diagnosed cancer and the number of diagnosis is growing worldwide due to increased exposure to solar radiation and the aging population. Reduction of positive margin rates when removing BCC leads to fewer revision surgeries and consequently lower health care costs, improved cosmetic outcomes and better patient care. In this study, we propose the first use of a perioperative mass spectrometry technology (iKnife) along with a deep learning framework for detection of BCC signatures from tissue burns.
Methods
Resected surgical specimen were collected and inspected by a pathologist. With their guidance, data were collected by burning regions of the specimen labeled as BCC or normal, with the iKnife. Data included 190 scans of which 127 were normal and 63 were BCC. A data augmentation approach was proposed by modifying the location and intensity of the peaks of the original spectra, through noise addition in the time and frequency domains. A symmetric autoencoder was built by simultaneously optimizing the spectral reconstruction error and the classification accuracy. Using t-SNE, the latent space was visualized.
Results
The autoencoder achieved an accuracy (standard deviation) of 96.62 (1.35%) when classifying BCC and normal scans, a statistically significant improvement over the baseline state-of-the-art approach used in the literature. The t-SNE plot of the latent space distinctly showed the separability between BCC and normal data, not visible with the original data. Augmented data resulted in significant improvements to the classification accuracy of the baseline model.
Conclusion
We demonstrate the utility of a deep learning framework applied to mass spectrometry data for surgical margin detection. We apply the proposed framework to an application with light surgical overhead and high incidence, the removal of BCC. The learnt models can accurately separate BCC from normal tissue.







Similar content being viewed by others
References
Canadian Cancer Society’s Advisory Committee on Cancer Statistics (2014) Canadian cancer statistics 2014. Canadian Cancer Society, Toronto
Moran MS, Schnitt SJ, Giuliano AE, Harris JR, Khan SA, Horton J, Klimberg S, Chavez-MacGregor M, Freedman G, Houssami N, Johnson PL, Morrow M (2014) Society of Surgical Oncology-American Society for Radiation Oncology consensus guideline on margins for breast-conserving surgery with whole-breast irradiation in stages I and II invasive breast cancer. Clin Oncol 10:1507–1515
Jung W, Kang E, Kim SM, Kim D, Hwang Y, Sun Y, Yom CK, Kim SW (2012) Factors associated with re-excision after breast-conserving surgery for early-stage breast cancer. J Breast Cancer 15:412–419
Balog J, Sasi-Szabo L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, Mirnezami R, Dezso B, Damjanovich L, Darzi A, Nicholson JK, Takats Z (2013) Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med 5(194):194
Strittmatter N, Jones EA, Veselkov KA, Rebec M, Bundy JG, Takats Z (2013) Analysis of intact bacteria using rapid evaporative ionisation mass spectrometry. Chem Commun 49:6188–6190
Agar NYR, Kowalski J, Kowalski PJ, Wong JH, Agar JN (2010) Tissue preparation for the in sity MALDI MS imaging of proteins, lipids, and small molecules at cellular resolution. Mass Spectrom Imaging 656:415–431
Verbeeck N, Caprioli RM, Van de Plas R (2018) Unsupervised machine learning for exploratory data analysis in imaging mass spectrometry. Mass Spectrom Rev 39:245–291. https://doi.org/10.1002/mas.21602
Thomas SA, Race AM, Steven RT (2016) Dimensionality reduction of mass spectrometry imaging data using autoencoders. In: IEEE symposium series on computational intelligence (SSCI), pp 1–7
Gut Y, Boiret M, Bultel L, Renaud T, Chetouani A, Hafiane A, Ginot YM, Jennane R (2015) Application of chemometric algorithms to MALDI mass spectrometry imaging of pharmaceutical tablets. J Pharm Biomed Anal 105:91–100
Behrmann J, Etmann C, Boskamp T, Casadonte R, Kriegsmann J, Maab P (2018) Deep learning for tumor classification in imaging mass spectrometry. Bioinformatics 34(7):1215–1223
John RE, Balog J, McKenzie JS, Rossi M, Covington A, Muihead L, Bodai Z, Rosini F, Speller AVM, Shousha S, Ramakrishnan R, Darzi A, Takats Z, Leff DR (2017) Rapid evaporative ionization mass spectrometry of electrosurgical vapours for identification of breast pathology: towards and intelligent knife for breast cancer surgery. Breast Cancer Res 19:1–14
Kinross JM, Muirhead L, Alexander J, Balog J, Guallar-Hoya C, Speller A, Golff O, Goldin R, Darzi A, Nicholson J, Takats Z (2016) iKnife: Rapid evaporative ionization mass spectrometry (REIMS) enables real-time chemical analysis of the mucosal lipidome for diagnostic and prognostic use in colorectal cancer. AACR 107th Annual Meeting. https://doi.org/10.1158/1538-7445.am2016-3977
Phelps DL, Balog J, Gildea LF, Bodai Z, Savage A, El-Bahrawy MA, Speller AV, Rosini F, Kudo H, McKenzie JS, Brown R, Takats Z, Ghaem-Maghami S (2018) The surgical intelligent knife distinguishes normal, borderline and malignant gynecological tissues using rapid evaporative ionization mass spectrometry (REIMS). Br J Cancer 118:1349–1358
Marcus D, Savage A, Balog J, Kudo H, Abda J, Dina R, Takats Z, Ghaem-Maghami (2019) Endometrial cancer: can the iKnife diagnose endometrial cancer? Int J Gynecol Cancer 29:A100–A101
Wang J, Perez L (2017) The effectiveness of data augmentation in image classification using deep learning. Technical Report: Preprint arXiv:1712.04621
Chen X, Duan Y, Houthooft R, Schulman J, Sutskever I, Abbeel P (2016) Info GAN: interpretable representation learning by information maximizing generativel adversarial nets. Advances in neural information processing systems, pp 2173–2180. Preprint arXiv:1606.03657
Maaten LVD, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9:2579–2605
Funding
This study was funded by The National Sciences and Engineering Research Council of Canada (NSERC), The Canadian Institutes of Health Research (CIHR) and Canada Foundation for Innovation (CFI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Ethical approval was not applicable since this study was ex vivo. Patients’ information was anonymized.
Informed consent
Statement of informed consent was not applicable since this study was ex vivo.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Santilli, A.M.L., Jamzad, A., Janssen, N.N.Y. et al. Perioperative margin detection in basal cell carcinoma using a deep learning framework: a feasibility study. Int J CARS 15, 887–896 (2020). https://doi.org/10.1007/s11548-020-02152-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-020-02152-9