Abstract
Key message
Successful orange rust development on sugarcane can potentially be explained as suppression of the plant immune system by the pathogen or delayed plant signaling to trigger defense responses.
Abstract
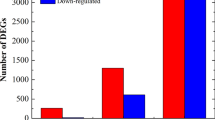
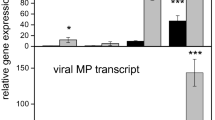
Puccinia kuehnii is an obligate biotrophic fungus that infects sugarcane leaves causing a disease called orange rust. It spread out to other countries resulting in reduction of crop yield since its first outbreak. One of the knowledge gaps of that pathosystem is to understand the molecular mechanisms altered in susceptible plants by this biotic stress. Here, we investigated the changes in temporal expression of transcripts in pathways associated with the immune system. To achieve this purpose, we used RNA-Seq to analyze infected leaf samples collected at five time points after inoculation. Differential expression analyses of adjacent time points revealed substantial changes at 12, 48 h after inoculation and 12 days after inoculation, coinciding with the events of spore germination, haustoria post-penetration and post-sporulation, respectively. During the first 24 h, a lack of transcripts involved with resistance mechanisms was revealed by underrepresentation of hypersensitive and defense response related genes. However, two days after inoculation, upregulation of genes involved with immune response regulation provided evidence of some potential defense response. Events related to biotic stress responses were predominantly downregulated in the initial time points, but expression was later restored to basal levels. Genes involved in carbohydrate metabolism showed evidence of repression followed by upregulation, possibly to ensure the pathogen nutritional requirements were met. Our results support the hypothesis that P. kuehnii initially suppressed sugarcane genes involved in plant defense systems. Late overexpression of specific regulatory pathways also suggests the possibility of an inefficient recognition system by a susceptible sugarcane genotype.



Similar content being viewed by others
References
Ahmed MM, Ji W, Wang M, Bian S, Xu M, Wang W, Zhang J, Xu Z, Yu M, Liu Q, Zhang C, Zhang H, Tang S, Gu M, Yu H (2017) Transcriptional changes of rice in response to rice black-streaked dwarf virus. Gene 628(July):38–47. https://doi.org/10.1016/j.gene.2017.07.015 http://linkinghub.elsevier.com/retrieve/pii/S0378111917305279
Ahn Ip, Kim S, Lee Yh (2005) Vitamin B 1 functions as an activator of plant disease resistance 1. Plant Physiol 138(July):1505–1515. https://doi.org/10.1104/pp.104.058693.cytosolic
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Balasubramanian V, Vashisht D, Cletus J, Sakthivel N (2012) Plant \(\beta\)-1,3-glucanases: their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol Lett 34(11):1983–1990. https://doi.org/10.1007/s10529-012-1012-6
Barbasso D, Jordão H, Maccheroni W, Boldini J, Bressiani J, Sanguino A (2010) First report of Puccinia kuehnii, causal agent of orange rust of sugarcane Brazil. Plant Disease 94(9):1170–1170. https://doi.org/10.1094/PDIS-94-9-1170C
Belarmino LC, Lane de Oliveira Silva R, da Mota Soares Cavalcanti N, Krezdorn N, Kido EA, Horres R, Winter P, Kahl G, Benko-Iseppon AM (2013) SymGRASS: a database of sugarcane orthologous genes involved in arbuscular mycorrhiza and root nodule symbiosis. BMC Bioinform 14(SUPPL.1):1–8. https://doi.org/10.1186/1471-2105-14-S1-S2
Berger S, Sinha AK, Roitsch T (2007) Plant physiology meets phytopathology: plant primary metabolism and plant pathogen interactions. J Exp Bot 58(15–16):4019–4026. https://doi.org/10.1093/jxb/erm298
Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, Delucia EH (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33(10):1597–1613. https://doi.org/10.1111/j.1365-3040.2010.02167.x
Birch PRJ, Kamoun S (2000) Studying interaction transcriptomes: coordinated analyses of gene expression during plant-microorganism interactions. New Technologies for Life Sciences: A Trends Guide 1931(December):77–82, (internal-pdf://046-2869556480/046.pdf)
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo TH, Davis FG, Bateman J, Bryant S, Guzikowski AR, Tsai SL, Coyne S, Ye WW, Freeman RM, Peshkin L, Tabin CJ, Regev A, Haas BJ, Whited JL (2017) A tissue-mapped Axolotl De Novo transcriptome enables identification of limb regeneration factors. Cell Rep 18(3):762–776. https://doi.org/10.1016/j.celrep.2016.12.063, http://linkinghub.elsevier.com/retrieve/pii/S2211124716317703
Cardoso-Silva CB, Costa EA, Mancini MC, Balsalobre TWA, Costa Canesin LE, Pinto LR, Carneiro MS, Garcia AAF, De Souza AP, Vicentini R (2014) De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS One 9(2), https://doi.org/10.1371/journal.pone.0088462
Carpio LGT, Simone de Souza F (2017) Optimal allocation of sugarcane bagasse for producing bioelectricity and second generation ethanol in Brazil: scenarios of cost reductions. Renew Energy 111:771–780. https://doi.org/10.1016/j.renene.2017.05.015
Chapola RG, Hoffmann HP, Massola NS (2016) Reaction of sugarcane varieties to orange rust (Puccinia kuehnii) and methods for rapid identification of resistant genotypes. Trop Plant Pathol 41(3):139–146. https://doi.org/10.1007/s40858-016-0076-6
Cheavegatti-Gianotto A, de Abreu HMC, Arruda P, Bespalhok Filho JC, Burnquist WL, Creste S, di Ciero L, Ferro JA, de Oliveira Figueira AV, de Sousa Filgueiras T, Grossi-de Sá MdF, Guzzo EC, Hoffmann HP, de Andrade Landell MG, Macedo N, Matsuoka S, de Castro Reinach F, Romano E, da Silva WJ, de Castro Silva Filho M, César Ulian E (2011) Sugarcane (Saccharum X officinarum): a reference study for the regulation of genetically modified cultivars in Brazil. Trop Plant Biol 4(1):62–89. https://doi.org/10.1007/s12042-011-9068-3
Cho WK, Lian S, Kim SM, Seo BY, Jung JK, Kim KH (2015) Time-course RNA-Seq analysis reveals transcriptional changes in rice plants triggered by rice stripe virus infection. PLoS One 10(8):1–20. https://doi.org/10.1371/journal.pone.0136736
Cia MC, de Carvalho G, Azevedo RA, Monteiro-Vitorello CB, Souza GM, Nishiyama-Junior MY, Lembke CG, de Faria Antunes, RSdC, Marques JPR, Melotto M, Camargo LEA, (2018) Novel Insights Into the Early Stages of Ratoon Stunting Disease of Sugarcane Inferred from Transcript and Protein Analysis. Phytopathology 108(12):1455–1466. https://doi.org/10.1094/PHYTO-04-18-0120-R
Comstock JC, Sood SG, Glynn NC, Shine JM, McKemy JM, Castlebury LA (2008) First report of Puccinia kuehnii, causal agent of orange rust of sugarcane, in the United States and Western Hemisphere. Plant Dis 92(1):175–175. https://doi.org/10.1094/PDIS-92-1-0175A
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Daniels J, Roach BT (1987) Taxonomy and evolution. In: Heinz DJ (ed) Developments in crop science, Developments in crop science, vol 11, Elsevier, pp 7–84, https://doi.org/10.1016/B978-0-444-42769-4.50007-2, http://www.sciencedirect.com/science/article/pii/B9780444427694500072, arXiv:1011.1669v3
De Araujo PG, Rossi M, De Jesus EM, Saccaro NL, Kajihara D, Massa R, De Felix JM, Drummond RD, Falco MC, Chabregas SM, Ulian EC, Menossi M, Sluys MAV (2005) Transcriptionally active transposable elements in recent hybrid sugarcane. Plant J 44(5):707–717. https://doi.org/10.1111/j.1365-313X.2005.02579.x
de Setta N, Monteiro-Vitorello C, Metcalfe C, Cruz GM, Del Bem L, Vicentini R, Nogueira FT, Campos R, Nunes S, Turrini PC, Vieira A, Ochoa Cruz E, Corrêa TC, Hotta C, de Mello Varani A, Vautrin S, da Trindade A, de Mendonça Vilela M, Lembke C, Sato P, de Andrade R, Nishiyama M, Cardoso-Silva C, Scortecci K, Garcia AA, Carneiro M, Kim C, Paterson AH, Bergès H, D’Hont A, de Souza A, Souza G, Vincentz M, Kitajima J, Van Sluys MA (2014) Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15(1):540. https://doi.org/10.1186/1471-2164-15-540
de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bölter B, Delfino L, Truman W, Mansfield J, Smirnoff N, Grant M (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plants 1(6):15074. https://doi.org/10.1038/nplants.2015.74
Demidchik V (2015) Mechanisms of oxidative stress in plants: from classical chemistry to cell biology. Environ Exp Bot 109:212–228. https://doi.org/10.1016/j.envexpbot.2014.06.021
Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I, Feussner K, Meinicke P, Stierhof YD, Schwarz H, Macek B, Mann M, Kahmann R (2011) Metabolic priming by a secreted fungal effector. Nature 478(7369):395–398. https://doi.org/10.1038/nature10454
dos Santos FR, Zucchi MI, Park JW, Benatti TR, da Silva JA, Souza GM, Landell MG, Pinto LR (2017) New sugarcane microsatellites and target region amplification polymorphism primers designed from candidate genes related to disease resistance. Sugar Tech 19(2):219–224. https://doi.org/10.1007/s12355-016-0457-7
Dobon A, Bunting DCE, Cabrera-Quio LE, Uauy C, Saunders DGO (2016) The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics 17(1):380. https://doi.org/10.1186/s12864-016-2684-4
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol10(4):366–371, https://doi.org/10.1016/j.pbi.2007.04.020, arXiv:1011.1669v3
Fang J, Lin A, Qiu W, Cai H, Umar M, Chen R, Ming R (2016) Transcriptome profiling revealed stress-induced and disease resistance genes up-regulated in PRSV resistant transgenic papaya. Front Plant Sci 7(June):1–20. https://doi.org/10.3389/fpls.2016.00855
Fesel PH, Zuccaro A (2016) \(\beta\)-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet Biol 90:53–60. https://doi.org/10.1016/j.fgb.2015.12.004, http://linkinghub.elsevier.com/retrieve/pii/S1087184515300529
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucl Acids Res 44(D1):D279–D285. https://doi.org/10.1093/nar/gkv1344
Funes C, Pérez Gómez SG, Henriquez DD, Di Pauli V, Bertani RP, Fontana DP, Rago AM, Joya CM, Sopena RA, González V, Babi H, Erazzu LE, Cuenya MI, Ploper LD (2016) First report of orange rust of sugarcane caused by Puccinia kuehnii in Argentina. Plant Dis 100(4):861. https://doi.org/10.1094/PDIS-09-15-1099-PDN
Galat A (2003) Peptidylprolyl Cis / Trans Isomerases (Immunophilins): Biological diversity—targets—functions. Curr Top Med Chem 3(12):1315–1347. https://doi.org/10.2174/1568026033451862, http://www.ncbi.nlm.nih.gov/pubmed/12871165 http://www.eurekaselect.com/openurl/content.php?genre=article&issn=1568-0266&volume=3&issue=12&spage=1315
Gervasi F, Ferrante P, Dettori MT, Scortichini M, Verde I (2018) Transcriptome reprogramming of resistant and susceptible peach genotypes during Xanthomonas arboricola pv. pruni early leaf infection. PLOS ONE 13(4):e0196590. https://doi.org/10.1371/journal.pone.0196590
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Ann Rev Phytopathol 43(1):205–227. https://doi.org/10.1146/annurev.phyto.43.040204.135923
Glynn NC, Dixon LJ, Castlebury LA, Szabo LJ, Comstock JC (2010) PCR assays for the sugarcane rust pathogens Puccinia kuehnii and P. melanocephala and detection of a SNP associated with geographical distribution in P. kuehnii. Plant Pathol 59(4):703–711. https://doi.org/10.1111/j.1365-3059.2010.02299.x
Glynn NC, Laborde C, Davidson RW, Irey MS, Glaz B, D’Hont A, Comstock JC (2013) Utilization of a major brown rust resistance gene in sugarcane breeding. Mol Breed 31(2):323–331. https://doi.org/10.1007/s11032-012-9792-x
Godoy AV, Lazzaro AS, Casalongué CA, San Segundo B (2000) Expression of a Solanum tuberosum cyclophilin gene is regulated by fungal infection and abiotic stress conditions. Plant Sci 152(2):123–134. https://doi.org/10.1016/S0168-9452(99)00211-3
Gomez SGP (2013) Quantificação de parâmetros monocíclicos da ferrugem alaranjada (Puccinia kuehnii) em cana-de-açúcar. PhD thesis, Universidade de São Paulo, Piracicaba, https://doi.org/10.11606/D.11.2013.tde-15052013-090620
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652. https://doi.org/10.1038/nbt.1883, http://www.nature.com/nbt/journal/v29/n7/abs/nbt.1883.html#supplementary-information
Grivet L, Arruda P (2002) Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr Opin Plant Biol 5(2):122–127. https://doi.org/10.1016/S1369-5266(02)00234-0
Grivet L, Daniels C, Glaszmann JC, Hont aD, D’Hont A (2004) A review of recent molecular genetics evidence for sugarcane evolution and domestication. Ethnobot Res Appl 2:9–17
Hammond-Kosack KE, Jones JDG (1996) Resistance gene-dependent plant defense responses. Plant Cell 8(10):1773. https://doi.org/10.2307/3870229
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Ann Rev Plant Physiol Plant Mol Biol 48(1):575–607. https://doi.org/10.1146/annurev.arplant.48.1.575, http://www.ncbi.nlm.nih.gov/pubmed/15012275
He X, Huo Y, Liu X, Zhou Q, Feng S, Shen X, Li B, Wu S, Chen X (2018) Activation of disease resistance against Botryosphaeria dothidea by downregulating the expression of MdSYP121 in apple. Hortic Res 5:24. https://doi.org/10.1038/s41438-018-0030-5
Hoang NV, Furtado A, Mason PJ, Marquardt A, Kasirajan L, Thirugnanasambandam PP, Botha FC, Henry RJ (2017) A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genomics 18(1):395. https://doi.org/10.1186/s12864-017-3757-8
Inzé D, Montagu MV (1995) Oxidative stress in plants. Curr Opin Biotechno 6(2):153–158. https://doi.org/10.1016/0958-1669(95)80024-7
Irvine JE (1999) Saccharum species as horticultural classes. Theor Appl Genet 98(2):186–194. https://doi.org/10.1007/s001220051057
Iskandar HM, Simpson RS, Casu RE, Bonnett GD, Maclean DJ, Manners JM (2004) Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol Biol Reporter 22(4):325–337. https://doi.org/10.1007/BF02772676
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329. https://doi.org/10.1038/nature05286
Kamarudin AN, Lai KS, Lamasudin DU, Idris AS, Balia Yusof ZN (2017) Enhancement of thiamine biosynthesis in oil palm seedlings by colonization of endophytic fungus Hendersonia toruloidea. Front Plant Sci 8(October):1–8. https://doi.org/10.3389/fpls.2017.01799
Kan J, Liu T, Ma N, Li H, Li X, Wang J, Zhang B, Chang Y, Lin J (2017) Transcriptome analysis of Callery pear (Pyrus calleryana) reveals a comprehensive signalling network in response to Alternaria alternata. PLOS One 12(9):e0184988. https://doi.org/10.1371/journal.pone.0184988
Kanzaki H, Saitoh H, Ito A, Fujisawa S, Kamoun S, Katou S, Yoshioka H, Terauchi R (2003) Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol Plant Pathol 4(5):383–391. https://doi.org/10.1046/j.1364-3703.2003.00186.x
Kazan K, Lyons R (2014) Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26(6):2285–2309. https://doi.org/10.1105/tpc.114.125419
Kim C, Wang X, Lee TH, Jakob K, Lee GJ, Paterson AH (2014) Comparative analysis of miscanthus and saccharum reveals a shared whole-genome duplication but different evolutionary fates. Plant Cell 26(6):2420–2429. https://doi.org/10.1105/tpc.114.125583
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. https://doi.org/10.1038/nmeth.3317, http://www.ncbi.nlm.nih.gov/pubmed/25751142%5Cnwww.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4655817, 15334406
Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416(6879):447–451. https://doi.org/10.1038/416447a
Kopylova E, Noé L, Touzet H (2012) SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28(24):3211–3217. https://doi.org/10.1093/bioinformatics/bts611
Lehmann S, Serrano M, L’Haridon F, Tjamos SE, Metraux JP (2015) Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112(1):54–62. https://doi.org/10.1016/j.phytochem.2014.08.027
Leme GM, Coutinho ID, Creste S, Hojo O, Carneiro RL, Bolzani VDS, Cavalheiro AJ (2014) HPLC-DAD method for metabolic fingerprinting of the phenotyping of sugarcane genotypes. Anal Methods 6(19):7781–7788. https://doi.org/10.1039/c4ay01750a
Li X, Xia B, Jiang Y, Wu Q, Wang C, He L, Peng F, Wang R (2010) A new pathogenesis-related protein, LrPR4, from Lycoris radiata, and its antifungal activity against Magnaporthe grisea. Molecular Biology Reports 37(2):995–1001. https://doi.org/10.1007/s11033-009-9783-0
Li ZY, Wang N, Dong L, Bai H, Quan JZ, Liu L, Dong ZP (2015) Differential gene expression in foxtail millet during incompatible interaction with Uromyces setariae-italicae. PLoS One 10(4):1–16. https://doi.org/10.1371/journal.pone.0123825
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930. https://doi.org/10.1093/bioinformatics/btt656
Liebthal M, Maynard D, Dietz KJ (2018) Peroxiredoxins and Redox Signaling in Plants. Antioxidants & Redox Signaling 28(7):609–624. https://doi.org/10.1089/ars.2017.7164
Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R (2015) Fungal effectors and plants susceptibility. Ann Rev Plant Biol 66(1):513–545. https://doi.org/10.1146/annurev-arplant-043014-114623
Loarce Y, Navas E, Paniagua C, Fominaya A, Manjón JL, Ferrer E (2016) Identification of genes in a partially resistant genotype of avena sativa expressed in response to Puccinia coronata infection. Front Plant Sci 7(May):1–15. https://doi.org/10.3389/fpls.2016.00731
Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B (2014) Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ 37(5):1250–1258. https://doi.org/10.1111/pce.12231
Lun AT, Chen Y, Smyth GK (2016) It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol Biol 1418:391–416. https://doi.org/10.1007/978-1-4939-3578-9_19
Lysøe E, Seong KY, Kistler HC (2011) The transcriptome of Fusarium graminearum during the infection of wheat. Mol Plant-Microbe Interactions 24(9):995–1000. https://doi.org/10.1094/mpmi-02-11-0038
Magarey R, Willcox T, Croft B, Cordingley A (2001) Orange Rust, A Major Pathogen Affecting Crops of Q124 in Queenland in 2000. Proc Aust Soc Sugarcane Technol 23:274–280
Manochio C, Andrade B, Rodriguez R, Moraes B (2017) Ethanol from biomass: a comparative overview. Renew Sustain Energy Rev 80(February):743–755. https://doi.org/10.1016/j.rser.2017.05.063, http://linkinghub.elsevier.com/retrieve/pii/S1364032117307001
Marques JPR, Hoy JW, Appezzato-da Glória B, Viveros AFG, Vieira MLC, Baisakh N (2018) Sugarcane cell wall-associated defense responses to infection by Sporisorium scitamineum. Front Plant Sci 9(May):1–14. https://doi.org/10.3389/fpls.2018.00698
Marra R, Ambrosino P, Carbone V, Vinale F, Woo SL, Ruocco M, Ciliento R, Lanzuise S, Ferraioli S, Soriente I, Gigante S, Turrà D, Fogliano V, Scala F, Lorito M (2006) Study of the three-way interaction between Trichoderma atroviride, plant and fungal pathogens by using a proteomic approach. Curr Genet 50(5):307–321. https://doi.org/10.1007/s00294-006-0091-0
Mattiello L, Riaño-pachón DM, Camara M, Martins M, Prado L, Bassi D, Eduardo P, Marchiori R, Ribeiro RV, Labate MTV, Labate CA, Menossi M (2015) Physiological and transcriptional analyses of developmental stages along sugarcane leaf. BMC Plant Biology pp 1–21, https://doi.org/10.1186/s12870-015-0694-z
McDowell JM, Woffenden BJ (2003) Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol 21(4):178–183. https://doi.org/10.1016/S0167-7799(03)00053-2
McNeil MD, Bhuiyan SA, Berkman PJ, Croft BJ, Aitken KS (2018) Analysis of the resistance mechanisms in sugarcane during Sporisorium scitamineum infection using RNA-seq and microscopy. PLOS ONE 13(5):e0197840. https://doi.org/10.1371/journal.pone.0197840
Medeiros CNF, Gonçalves MC, Harakava R, Creste S, Nóbile PM, Pinto LR, Perecin D, Landell MGA (2014) Sugarcane transcript profiling assessed by cDNA-AFLP analysis during the interaction with Sugarcane Mosaic Virus (July):511–520
Melayah D, Bonnivard E, Chalhoub B, Audeon C, Grandbastien MA (2001) The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J 28(2):159–168. https://doi.org/10.1046/j.1365-313X.2001.01141.x
Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: The new wave? Trends Plant Sci 16(6):300–309. https://doi.org/10.1016/j.tplants.2011.03.007
Moraes BS, Zaiat M, Bonomi A (2015) Anaerobic digestion of vinasse from sugarcane ethanol production in Brazil: challenges and perspectives. Renew Sustain Energy Rev 44:888–903. https://doi.org/10.1016/j.rser.2015.01.023
Moreira AS, Nogueira Junior AF, Gonçalves CRNB, Souza NA, Bergamin Filho A (2018) Pathogenic and molecular comparison of Puccinia kuehnii isolates and reactions of sugarcane varieties to orange rust. Plant Pathol I:1–10. https://doi.org/10.1111/ppa.12870
Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA (2010) Genome-wide Anal Phenylpropanoid Defence Pathways. https://doi.org/10.1111/j.1364-3703.2010.00648.x
Nejat N, Cahill DM, Vadamalai G, Ziemann M, Rookes J, Naderali N (2015) Transcriptomics-based analysis using RNA-Seq of the coconut (Cocos nucifera) leaf in response to yellow decline phytoplasma infection. Molecular Genetics and Genomics 290(5):1899–1910. https://doi.org/10.1007/s00438-015-1046-2
Nicholson RL, Hammerschmidt R (1992) Phenolic compounds and their role in disease resistance. Ann Rev Phytopathol 30(1):369–389. https://doi.org/10.1146/annurev.py.30.090192.002101
O’Connell RJ, Panstruga R (2006) Tete a tete inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol 171(4):699–718. https://doi.org/10.1111/j.1469-8137.2006.01829.x
Páez UAH, Romero IAG, Restrepo SR, Gutiérrez FAA, Castaño DM (2015) Assembly and analysis of differential transcriptome responses of Hevea brasiliensis on interaction with Microcyclus ulei. PLoS One 10(8):1–21. https://doi.org/10.1371/journal.pone.0134837
Pandey S, Somssich IE (2009) The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiology 150(August):1648–1655. https://doi.org/10.1104/pp.109.138990
Parco AS, Hale AL, Avellaneda MC, Hoy JW, Kimbeng CA, Pontif MJ, McCord PH, Ayala-Silva T, Todd JR, Baisakh N (2017) Distribution and frequency of Bru1, a major brown rust resistance gene, in the sugarcane world collection. Plant Breed 136(5):637–651. https://doi.org/10.1111/pbr.12508
Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24(5):255–265, https://doi.org/10.1007/s00299-005-0972-6, arXiv:1011.1669v3
Paterson AH, Moore PH, Tew TL (2013) The gene pool of Saccharum species and their improvement, Springer New York, New York, NY, pp 43–71. https://doi.org/10.1007/978-1-4419-5947-8_3
Patkar RN, Naqvi NI (2017) Fungal manipulation of hormone-regulated plant defense. PLOS Pathogens 13(6):e1006334. https://doi.org/10.1371/journal.ppat.1006334
Peters LP, Carvalho G, Vilhena MB, Creste S, Azevedo RA, Monteiro-Vitorello CB (2017) Functional analysis of oxidative burst in sugarcane smut-resistant and -susceptible genotypes. Planta 245(4):749–764. https://doi.org/10.1007/s00425-016-2642-z
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30(9):e36, http://www.ncbi.nlm.nih.gov/pubmed/11972351 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC113859
Piperidis G, Piperidis N, D’Hont A (2010) Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genomics 284(1):65–73. https://doi.org/10.1007/s00438-010-0546-3
Piterková J, Luhová L, Mieslerová B, Lebeda A, Petřivalský M (2013) Nitric oxide and reactive oxygen species regulate the accumulation of heat shock proteins in tomato leaves in response to heat shock and pathogen infection. Plant Sci 207:57–65. https://doi.org/10.1016/j.plantsci.2013.02.010, http://linkinghub.elsevier.com/retrieve/pii/S0168945213000411
Que Y, Su Y, Guo J, Wu Q, Xu L (2014) A global view of transcriptome dynamics during Sporisorium scitamineum challenge in sugarcane by RNA-seq. PLoS One 9(8):e106476. https://doi.org/10.1371/journal.pone.0106476
Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339(1):62–6, http://www.ncbi.nlm.nih.gov/pubmed/12618301
Reddy VS, Ali GS, Reddy ASN (2003) Characterization of a pathogen-induced calmodulin-binding protein: mapping of four Ca2+-dependent calmodulin-binding domains. Plant Mol Biol 52(1):143–159. https://doi.org/10.1023/A:1023993713849
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Rodrigues Reis CE, Hu B (2017) Vinasse from sugarcane ethanol production: Better treatment or better utilization? Front Energy Res 5(April):1–7. https://doi.org/10.3389/fenrg.2017.00007
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends in Plant Science 15(5):247–258. https://doi.org/10.1016/j.tplants.2010.02.006, http://linkinghub.elsevier.com/retrieve/pii/S1360138510000324
Santa Brigida AB, Rojas CA, Grativol C, De Armas EM, Entenza JO, Thiebaut F, Lima MDF, Farrinelli L, Hemerly AS, Lifschitz S, Ferreira PC (2016) Sugarcane transcriptome analysis in response to infection caused by Acidovorax avenae subsp. avenae. PLoS One 11(12):1–30, https://doi.org/10.1371/journal.pone.0166473
Schaker PDC, Palhares AC, Taniguti LM, Peters LP, Creste S, Aitken KS, Van Sluys MA, Kitajima JP, Vieira MLC, Monteiro-Vitorello CB (2016) RNAseq Transcriptional Profiling following Whip Development in Sugarcane Smut Disease. PLOS ONE 11(9):e0162237. https://doi.org/10.1371/journal.pone.0162237
Schaker PDC, Peters LP, Cataldi TR, Labate CA, Caldana C, Monteiro-Vitorello CB (2017) Metabolome dynamics of smutted sugarcane reveals mechanisms involved in disease progression and whip emission. Front Plant Sci 8(May):1–17. https://doi.org/10.3389/fpls.2017.00882
Socquet-Juglard D, Kamber T, Pothier JF, Christen D, Gessler C, Duffy B, Patocchi A (2013) Comparative RNA-seq analysis of early-infected peach leaves by the invasive phytopathogen Xanthomonas arboricola pv. pruni. PLoS One 8(1):e54196. https://doi.org/10.1371/journal.pone.0054196
Spanu PD, Panstruga R (2017) Editorial: Biotrophic Plant-Microbe Interactions. Frontiers in Plant Science 8(February):1–4. https://doi.org/10.3389/fpls.2017.00192
Su Y, Xu L, Wang S, Wang Z, Yang Y, Chen Y, Que Y (2015) Identification, phylogeny, and transcript of chitinase family genes in sugarcane. Sci Rep 5:1–15. https://doi.org/10.1038/srep10708
Svensson B, Svendsen I, Hoejrup P, Roepstorff P, Ludvigsen S, Poulsen FM (1992) Primary structure of barwin: a barley seed protein closely related to the C-terminal domain of proteins encoded by wound-induced plant genes. Biochemistry 31(37):8767–8770. https://doi.org/10.1021/bi00152a012
Takabatake R, Karita E, Seo S, Mitsuhara I, Kuchitsu K, Ohashi Y (2007) Pathogen-induced calmodulin isoforms in Basal resistance against bacterial and fungal pathogens in Tobacco. Plant Cell Physio 48(3):414–423. https://doi.org/10.1093/pcp/pcm011
Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151(2):936–948. https://doi.org/10.1104/pp.109.145623
Teixeira PJPL, Thomazella DPdT, Reis O, do Prado PFV, do Rio MCS, Fiorin GL, José J, Costa GGL, Negri VA, Mondego JMC, Mieczkowski P, Pereira GAG, (2014) High-resolution transcript profiling of the atypical biotrophic interaction between Theobroma cacao and the fungal pathogen Moniliophthora perniciosa. Plant Cell Online 26(11):4245–4269. https://doi.org/10.1105/tpc.114.130807
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37(6):914–939. https://doi.org/10.1111/j.1365-313X.2004.02016.x
Thokoane L, Rutherford R (2001) cDNA-AFLP differential display of sugarcane (Saccharum spp, hybrids) genes induced by challenge with the fungal pathogen Ustilago scitaminea (sugarcane smut). Proc S Afr Sug Technol Ass 75(m):104–107
Tremblay A, Hosseini P, Li S, Alkharouf NW, Matthews BF (2013) Analysis of Phakopsora pachyrhizi transcript abundance in critical pathways at four time-points during infection of a susceptible soybean cultivar using deep sequencing. BMC Genomics 14(1):614. https://doi.org/10.1186/1471-2164-14-614
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3-new capabilities and interfaces. Nucl Acids Res 40(15):e115–e115. https://doi.org/10.1093/nar/gks596
Vasudevan D, Gopalan G, Kumar A, Garcia VJ, Luan S (1850) Swaminathan K (2015) plant immunophilins: a review of their structure-function relationship. Biochimica et Biophys Acta Gen Sub 10:2145–2158. https://doi.org/10.1016/j.bbagen.2014.12.017
Vicentini R, Bottcher A, Dos Santos Brito M, Dos Santos AB, Creste S, De Andrade Landell MG, Cesarino I, Mazzafera P (2015) Large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PLoS One 10(8):e0134909. https://doi.org/10.1371/journal.pone.0134909
Voegele RT, Mendgen K (2003) Rust haustoria: nutrient uptake and beyond. New Phytologist 159(1):93–100. https://doi.org/10.1046/j.1469-8137.2003.00761.x
Wang L, Ye Liu H, Liu X, Wei C, Huang Y, Liu Y, Tu J (2016) Both overexpression and suppression of an Oryza sativa NB-LRR-like gene OsLSR result in autoactivation of immune response and thiamine accumulation. Sci Rep 6(1):24079. https://doi.org/10.1038/srep24079
Westermann AJ, Gorski SA, Vogel J (2012) Dual RNA-seq of pathogen and host. Nat Rev Microbiol 10(9):618–630. https://doi.org/10.1038/nrmicro2852
Xia C, Li S, Hou W, Fan Z, Xiao H, Lu M, Sano T, Zhang Z (2017) Global transcriptomic changes induced by infection of cucumber (Cucumis sativus L.) with mild and severe variants of hop stunt viroid. Front Microbiol 8(DEC):1–16. https://doi.org/10.3389/fmicb.2017.02427
Yadav IS, Sharma A, Kaur S, Nahar N, Bhardwaj SC, Sharma TR, Chhuneja P (2016) Comparative temporal transcriptome profiling of wheat near isogenic line carrying Lr57 under compatible and incompatible interactions. Front Plant Sci 7:1943. https://doi.org/10.3389/fpls.2016.01943
Yang X, Sood S, Glynn N, Islam MS, Comstock J, Wang J (2017) Constructing high-density genetic maps for polyploid sugarcane (Saccharum spp.) and identifying quantitative trait loci controlling brown rust resistance. Mol Breed 37(10), https://doi.org/10.1007/s11032-017-0716-7
Yang X, Islam MS, Sood S, Maya S, Hanson EA, Comstock J, Wang J (2018) Identifying quantitative trait Loci (QTLs) and developing diagnostic markers linked to orange rust resistance in sugarcane (SaccharumA spp.). Front Plant Sci 9(March):1–10. https://doi.org/10.3389/fpls.2018.00350
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14. https://doi.org/10.1186/gb-2010-11-2-r14
Zhang H, Yang Y, Wang C, Liu M, Li H, Fu Y, Wang Y, Nie Y, Liu X, Ji W (2014) Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics 15(1):1–14. https://doi.org/10.1186/1471-2164-15-898
Zhang J, Nagai C, Yu Q, Pan YB, Ayala-Silva T, Schnell RJ, Comstock JC, Arumuganathan AK, Ming R (2012) Genome size variation in three Saccharum species. Euphytica 185(3):511–519. https://doi.org/10.1007/s10681-012-0664-6
Zhang Y, Tian L, Yan DH, He W (2018) Genome-wide transcriptome analysis reveals the comprehensive response of two susceptible poplar sections to Marssonina brunnea infection. Genes 9(3):154. https://doi.org/10.3390/genes9030154
Zhao D, Glynn NC, Glaz B, Comstock JC, Sood S (2011) Orange Rust Effects on Leaf Photosynthesis and Related Characters of Sugarcane. Plant Disease 95(6):640–647. https://doi.org/10.1094/PDIS-10-10-0762
Zhu L, Ni W, Liu S, Cai B, Xing H, Wang S (2017) Transcriptomics analysis of apple leaves in response to Alternaria alternata apple pathotype infection. Front Plant Sci 8:22. https://doi.org/10.3389/fpls.2017.00022
Acknowledgements
FHC received a fellowship grant #2015/17935-8, São Paulo Research Foundation (FAPESP). GRAM received a research grant #2015/22993-7, São Paulo Research Foundation (FAPESP). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001. This research was supported by the Brazilian Innovation Agency (FINEP), Grant number 01.09.0335.00 to MSC.
Author information
Authors and Affiliations
Contributions
SGPG, NSM and MSC designed the study. SGPG performed the phytopathology experiment. NSM, MSC, LEAC and GRAM planned the RNA sequencing and qPCR experiments. FHC and GKH performed the RNA extraction, and MCC performed the qPCR experiments. FHC, GKH and GRAM analyzed the RNA-Seq data. FHC, GKH, MCC, CBMV, LEAC and GRAM wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Prakash Lakshmanan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Correr, F.H., Hosaka, G.K., Gómez, S.G.P. et al. Time-series expression profiling of sugarcane leaves infected with Puccinia kuehnii reveals an ineffective defense system leading to susceptibility. Plant Cell Rep 39, 873–889 (2020). https://doi.org/10.1007/s00299-020-02536-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02536-w