Abstract
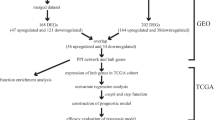
Hepatocellular carcinoma (HCC) is a common cancer of high mortality, mainly due to the difficulty in diagnosis during its clinical stage. Here we aim to find the gene biomarkers, which are of important significance for diagnosis and treatment. In this work, 3682 differentially expressed genes on HCC were firstly differentiated based on the Cancer Genome Atlas database (TCGA). Co-expression modules of these differentially expressed genes were then constructed based on the weighted correlation network algorithm. The correlation coefficient between the co-expression module and clinical data from the Broad GDAC Firehose was thereafter derived. Finally, the interactive network of genes was then constructed. Then, the hub genes were used to implement enrichment analysis and pathway analysis in the Database for Annotation, Visualization and Integrated Discovery (DAVID) database. Results revealed that the abnormally expressed genes in the module played an important role in the biological process including cell division, sister chromatid cohesion, DNA repair, and G1/S transition of mitotic cell cycle. Meanwhile, these genes also enriched in a few crucial pathways related to Cell cycle, Oocyte meiosis, and p53 signaling. Via investigating the closeness centrality of the interactive network, eight gene biomarkers including the CKAP2, TPX2, CDCA8, KIFC1, MELK, SGO1, RACGAP1, and KIAA1524 gene were discovered, whose functions had been indeed revealed to be correlated with HCC. This study, therefore, suggests that the abnormal expression of those eight genes may be taken as gene biomarkers of HCC.





Similar content being viewed by others
References
Han LL, Lv Y, Guo H et al (2014) Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J Gastroenterol 20(30):10249–10261. https://doi.org/10.3748/wjg.v20.i30.10249
Kim JU, Cox IJ, Taylor-Robinson SD (2017) The quest for relevant hepatocellular carcinoma biomarkers. Cell Mol Gastroenterol Hepatol 4(2):283–284. https://doi.org/10.1016/j.jcmgh.2017.06.003
Miura T, Ban D, Tanaka S et al (2015) Distinct clinicopathological phenotype of hepatocellular carcinoma with ethoxybenzyl-magnetic resonance imaging hyperintensity: association with gene expression signature. Am J Surg 210(3):561–569. https://doi.org/10.1016/j.amjsurg.2015.03.027
Zhou C, Zhang W, Chen W et al (2017) Integrated analysis of copy number variations and gene expression profiling in hepatocellular carcinoma. Sci Rep 7(1):10570. https://doi.org/10.1038/s41598-017-11029-y
Wei L, Lian B, Zhang Y et al (2014) Application of microRNA and mRNA expression profiling on prognostic biomarker discovery for hepatocellular carcinoma. BMC Genomics 15(1):S13. https://doi.org/10.1186/1471-2164-15-S1-S13
Maher CA, Kumar-Sinha C, Cao XH et al (2009) Transcriptome sequencing to detect gene fusions in cancer. Nature 458:97–101. https://doi.org/10.1038/nature07638
Griffith M, Griffith OL, Mwenifumbo J et al (2010) Alternative expression analysis by RNA sequencing. Nat Methods 7:843–847. https://doi.org/10.1038/nmeth.1503
Maher CA, Palanisamy N, Brenner JC et al (2009) Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci 106(30):12353–12358. https://doi.org/10.1073/pnas.0904720106
McCarthy DJ, Chen Y, Smyth GK et al (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40(10):4288–4297. https://doi.org/10.1093/nar/gks042
Zhao Q, Caballero OL, Levy S et al (2009) Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci 106(6):1886–1891. https://doi.org/10.1073/pnas.0812945106
Sinicropi D, Qu K, Collin F et al (2012) Whole transcriptome RNA-Seq analysis of breast cancer recurrence risk using formalin-fixed paraffin-embedded tumor tissue. PLoS ONE 7(7):e40092. https://doi.org/10.1371/journal.pone.0040092
Ren S, Peng Z, Mao JH et al (2012) RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res 22:806–821. https://doi.org/10.1038/cr.2012.30
Shah SP, Köbel M, Senz J et al (2009) Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med 360(26):2719–2729. https://doi.org/10.1056/NEJMoa0902542
Beane J, Vick J, Schembri F et al (2011) Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res 4(6):803–817. https://doi.org/10.1158/1940-6207.CAPR-11-0212
Liu J, Yu Z, Sun M et al (2019) Identification of cancer/testis antigen 2 gene as a potential hepatocellular carcinoma therapeutic target by hub gene screening with topological analysis. Oncol Lett 18(5):4778–4788. https://doi.org/10.3892/ol.2019.10811
Guo Y, Bao Y, Ma M et al (2017) Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int J Mol Sci 18(4):722. https://doi.org/10.3390/ijms18040722
Agarwal R, Narayan J, Bhattacharyya A et al (2017) Gene expression profiling, pathway analysis and subtype classification reveal molecular heterogeneity in hepatocellular carcinoma and suggest subtype specific therapeutic targets. Cancer Genetics 216:37–51. https://doi.org/10.1016/j.cancergen.2017.06.002
Chen L, Liu R, Liu ZP et al (2012) Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci Rep 2:342. https://doi.org/10.1038/srep00342
Li Y, Vongsangnak W, Chen L et al (2014) Integrative analysis reveals disease-associated genes and biomarkers for prostate cancer progression. BMC Med Genomics 7(1):S3. https://doi.org/10.1186/1755-8794-7-S1-S3
Yuan X, Chen J, Lin Y et al (2017) Network biomarkers constructed from gene expression and protein-protein interaction data for accurate prediction of leukemia. J Cancer 8(2):278–286. https://doi.org/10.7150/jca.17302
Solé X, Crous-Bou M, Cordero D et al (2014) Discovery and validation of new potential biomarkers for early detection of colon cancer. PLoS ONE 9(9):e106748. https://doi.org/10.1371/journal.pone.0106748
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. https://doi.org/10.2202/1544-6115.1128
Ravasz E, Somera AL, Mongru DA et al (2002) Hierarchical organization of modularity in metabolic networks. Science 297(5586):1551–1555. https://doi.org/10.1126/science.1073374
Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1(1):54. https://doi.org/10.1186/1752-0509-1-54
Yoo S, Wang W, Wang Q et al (2017) A pilot systematic genomic comparison of recurrence risks of hepatitis B virus-associated hepatocellular carcinoma with low- and high-degree liver fibrosis. BMC Med 15:214. https://doi.org/10.1186/s12916-017-0973-7
Wang SM, Ooi LL, Hui KM (2007) Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res 13(21):6275–6283. https://doi.org/10.1158/1078-0432.CCR-06-2236
Jiang Y, Sun A, Zhao Y et al (2019) Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 567:257–261. https://doi.org/10.1038/s41586-019-0987-8
Marte B (2004) Cell division and cancer. Nature 432:293. https://doi.org/10.1038/432293a
Liu C, Liu L, Chen X et al (2016) Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 64:117–129. https://doi.org/10.1002/hep.28509
Liu Q, Yang P, Tu K et al (2014) TPX2 knockdown suppressed hepatocellular carcinoma cell invasion via inactivating AKT signaling and inhibiting MMP2 and MMP9 expression. Chin J Cancer Res 26(4):410–417. https://doi.org/10.3978/j.issn.1000-9604.2014.08.01
Wang LH, Yen CJ, Li TN et al (2015) Sgo1 is a potential therapeutic target for hepatocellular carcinoma. Oncotarget 6(4):2023–2033. https://doi.org/10.18632/oncotarget.2764
Gramantieri L, Trerè D, Chieco P et al (2003) In human hepatocellular carcinoma in cirrhosis proliferating cell nuclear antigen (PCNA) is involved in cell proliferation and cooperates with P21 in DNA repair. J Hepatol 39(6):997–1003. https://doi.org/10.1016/S0168-8278(03)00458-6
Lukish JR, Muro K, DeNobile J et al (1998) Prognostic significance of DNA replication errors in young patients with colorectal cancer. Ann Surg 227(1):51–56. https://doi.org/10.1097/00000658-199801000-00008
Martin-Lluesma S, Schaeffer C, Robert EI et al (2008) Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology 48:1467–1476. https://doi.org/10.1002/hep.22542
Wu BK, Li CC, Chen HJ et al (2006) Blocking of G1/S transition and cell death in the regenerating liver of Hepatitis B virus X protein transgenic mice. Biochem Biophys Res Commun 340(3):916–928. https://doi.org/10.1016/j.bbrc.2005.12.089
Zhang L, Guo Y, Li B et al (2013) Identification of biomarkers for hepatocellular carcinoma using network-based bioinformatics methods. Eur J Med Res 18(1):35. https://doi.org/10.1186/2047-783X-18-35
Malumbres M, Barbacid M (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9:153–166. https://doi.org/10.1038/nrc2602
ElHefnawi M, Soliman B, Abu-Shahba N et al (2013) An integrative meta-analysis of microRNAs in hepatocellular carcinoma. Genom Proteom Bioinf 11(6):354–367. https://doi.org/10.1016/j.gpb.2013.05.007
Wong YH, Wu CC, Lin CL et al (2015) Applying NGS data to find evolutionary network biomarkers from the early and late stages of hepatocellular carcinoma. Biomed Res Int 2015:391475. https://doi.org/10.1155/2015/391475
Hucl T (2011) Gallmeier E (2011) DNA repair: exploiting the Fanconi anemia pathway as a potential therapeutic target. Physiol Res 60(3):453–465. https://doi.org/10.33549/physiolres.932115
Palagyi A, Neveling K, Plinninger U et al (2010) Genetic inactivation of the Fanconi anemia gene FANCC identified in the hepatocellular carcinoma cell line HuH-7 confers sensitivity towards DNA-interstrand crosslinking agents. Mol Cancer 9:127. https://doi.org/10.1186/1476-4598-9-127
Okayama A, Maruyama T, Tachibana N et al (1995) Increased prevalence of HTLV-I infection in patients with hepatocellular carcinoma associated with Hepatitis C virus. Cancer Sci 86:1–4. https://doi.org/10.1111/j.1349-7006.1995.tb02979.x
Lips EH, Mulder L, Hannemann J et al (2010) Indicators of homologous recombination deficiency in breast cancer and association with response to neoadjuvant chemotherapy. Ann Oncol 22(4):870–876. https://doi.org/10.1093/annonc/mdq468
Abkevich V, Timms KM, Hennessy BT et al (2012) Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 107:1776–1782. https://doi.org/10.1038/bjc.2012.451
Helleday T (2010) Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 31(6):955–960. https://doi.org/10.1093/carcin/bgq064
Kiss A, Wang NJ, Xie JP et al (1997) Analysis of transforming growth factor (TGF)-alpha/epidermal growth factor receptor, hepatocyte growth Factor/c-met, TGF-beta receptor type II, and p53 expression in human hepatocellular carcinomas. Clin Cancer Res 3(7):1059–1066
Greer EL, Brunet A (2005) FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24(50):7410–7425. https://doi.org/10.1038/sj.onc.1209086
Guo Q, Song Y, Hua K et al (2017) Involvement of FAK-ERK2 signaling pathway in CKAP2-induced proliferation and motility in cervical carcinoma cell lines. Sci Rep 7(1):2117. https://doi.org/10.1038/s41598-017-01832-y
Hayashi T, Ohtsuka M, Okamura D et al (2014) Cytoskeleton-associated protein 2 is a potential predictive marker for risk of early and extensive recurrence of hepatocellular carcinoma after operative resection. Surgery 155(1):114–123. https://doi.org/10.1016/j.surg.2013.06.009
Jeon TW, Min JK, Seo YR et al (2017) Abstract 3113: Knockdown of cell division cycle-associated 8 (CDCA8) suppresses hepatocellular carcinoma growth via the upregulation of tumor suppressor ATF3. Can Res 77(13):3113–3113. https://doi.org/10.1158/1538-7445.AM2017-3113
Fu X, Zhu Y, Zheng B et al (2018) KIFC1, a novel potential prognostic factor and therapeutic target in hepatocellular carcinoma. Int J Oncol 52(6):1912–1922. https://doi.org/10.3892/ijo.2018.4348
Xia H, Kong SN, Chen J et al (2016) MELK is an oncogenic kinase essential for early hepatocellular carcinoma recurrence. Cancer Lett 383(1):85–93. https://doi.org/10.1016/j.canlet.2016.09.017
Li Y, Li Y, Chen Y et al (2017) MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell Int 17(1):102. https://doi.org/10.1186/s12935-017-0471-1
Li Y, Zheng J, Yao J et al (2017) Aberrant expression and prognostic value of RacGAP1 in hepatocellular carcinoma. Int J Clin Exp Pathol 10:1747–1755
Junttila MR, Puustinen P, Niemelä M et al (2007) CIP2A inhibits PP2A in human malignancies. Cell 130(1):51–62. https://doi.org/10.1016/j.cell.2007.04.044
Chen KF, Liu CY, Lin YC et al (2010) CIP2A mediates effects of bortezomib on phospho-Akt and apoptosis in hepatocellular carcinoma cells. Oncogene 29:6257–6266. https://doi.org/10.1038/onc.2010.357
Côme C, Laine A, Chanrion M et al (2009) CIP2A is associated with human breast cancer aggressivity. Clin Cancer Res 15(16):5092–5100. https://doi.org/10.1158/1078-0432.CCR-08-3283
Li W, Ge Z, Liu C et al (2008) CIP2A is overexpressed in gastric cancer and its depletion leads to impaired clonogenicity, senescence, or differentiation of tumor cells. Clin Cancer Res 14(12):3722–3728. https://doi.org/10.1158/1078-0432.CCR-07-4137
Khanna A, Böckelman C, Hemmes A et al (2009) MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J Natl Cancer Inst 101(11):793–805. https://doi.org/10.1093/jnci/djp103
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grand Nos. 11371174 and 11804123) and Postgraduate Research and Practice Innovation Program of Jiangsu Province (No. KYCX18_1866). This manuscript had been accepted for presentation in CBC2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interest in this paper.
Rights and permissions
About this article
Cite this article
Qi, J., Zhou, J., Tang, XQ. et al. Gene Biomarkers Derived from Clinical Data of Hepatocellular Carcinoma. Interdiscip Sci Comput Life Sci 12, 226–236 (2020). https://doi.org/10.1007/s12539-020-00366-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-020-00366-8