Abstract
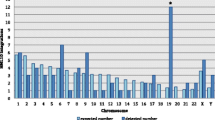
Increasing evidence suggests that DNA methylation has key roles in the replication of retroviruses, including lentiviruses, and pathogenesis of diseases. However, the precise characteristics of CpG islands are not known for many retroviruses. In this study, we compared the distribution of CpG islands among strains of equine infectious anemia virus (EIAV), a lentivirus in the family Retroviridae and a model for HIV research. We identified CpG islands in 32 full-length EIAV genomic sequences obtained from the GenBank database using MethPrimer. Only one CpG island, from 100 to 120 bp, was identified in the genomes of EIAV strains DV10, DLV3-A, and DLV5-10 from China, V26 and V70 from Japan, and IRE H3, IRE F2, IRE F3, and IRE F4 from Ireland. Importantly, the CpG island was located within the Rev gene, which is required for the expression of viral cis-acting elements and the production of new virions. These results suggest that the distribution, length, and genetic properties of CpG islands differ among EIAV strains. Future research should focus on the biological significance of this CpG island within rev to improve our understanding of the precise roles of CpG islands in epigenetic regulation in the species.



Similar content being viewed by others
References
Acevedo A, Brodsky L, Andino R (2014) Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505(7485):686–690
Alexandersen S, Carpenter S (1991) Characterization of variable regions in the envelope and S3 open reading frame of equine infectious anemia virus. J Virol 65(8):4255–4262
Ando M, Saito Y, Xu G, Bui NQ, Medetgul-Ernar K, Pu M, Fisch K, Ren S, Sakai A, Fukusumi T, Liu C, Haft S, Pang J, Mark A, Gaykalova DA, Guo T, Favorov AV, Yegnasubramanian S, Fertig EJ, Ha P, Tamayo P, Yamasoba T, Ideker T, Messer K, Califano JA (2019) Chromatin dysregulation and DNA methylation at transcription start sites associated with transcriptional repression in cancers. Nat Commun 10(1):2188
Belshan M, Baccam P, Oaks JL, Sponseller BA, Murphy SC, Cornette J, Carpenter S (2001) Genetic and biological variation in equine infectious anemia virus Rev correlates with variable stages of clinical disease in an experimentally infected pony. Virology 279(1):185–200
Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I (2009) CpG methylation controls reactivation of HIV from latency. PLoS Pathog 5(8):e1000554
Carpenter S, Chen W, Dorman K (2011) Rev Variation during Persistent Lentivirus Infection. Viruses 3(1):1–11
Carpenter S, Dobbs D (2010) Molecular and biological characterization of equine infectious anemia virus Rev. Curr HIV Res 8(1):87–93
Chavez L, Kauder S, Verdin E (2011) In vivo, in vitro, and in silico analysis of methylation of the HIV-1 provirus. Methods 53(1):47–53
Cook RF, Leroux C, Issel CJ (2013) Equine infectious anemia and equine infectious anemia virus in 2013: a review. Vet Microbiol 167(1–2):181–204
Craigo JK, Montelaro RC (2013) Lessons in AIDS vaccine development learned from studies of equine infectious, anemia virus infection and immunity. Viruses 5(12):2963–2976
Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25(10):1010–1022
Dong JB, Zhu W, Cook FR, Goto Y, Horii Y, Haga T (2013) Identification of a novel equine infectious anemia virus field strain isolated from feral horses in southern Japan. J Gen Virol 94(Pt 2):360–365
Dostal V, Churchill MEA (2019) Cytosine methylation of mitochondrial DNA at CpG sequences impacts transcription factor A DNA binding and transcription. Biochim Biophys Acta Gene Regul Mech 1862(5):598–607
Duverger A, Jones J, May J, Bibollet-Ruche F, Wagner FA, Cron RQ, Kutsch O (2009) Determinants of the establishment of human immunodeficiency virus type 1 latency. J Virol 83(7):3078–3093
Gallastegui E, Millan-Zambrano G, Terme JM, Chavez S, Jordan A (2011) Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J Virol 85(7):3187–3202
Han Y, Lin YB, An W, Xu J, Yang HC, O'Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF (2008) Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4(2):134–146
Hejnar J, Hajkova P, Plachy J, Elleder D, Stepanets V, Svoboda J (2001) CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc Natl Acad Sci USA 98(2):565–569
Hendrich B, Bird A (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 18(11):6538–6547
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492
Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19(2):187–191
Jordan A, Defechereux P, Verdin E (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20(7):1726–1738
Koiwa T, Hamano-Usami A, Ishida T, Okayama A, Yamaguchi K, Kamihira S, Watanabe T (2002) 5'-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J Virol 76(18):9389–9397
Lauring AS, Frydman J, Andino R (2013) The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 11(5):327–336
Lavie L, Kitova M, Maldener E, Meese E, Mayer J (2005) CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J Virol 79(2):876–883
Lenasi T, Contreras X, Peterlin BM (2008) Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4(2):123–133
Leroux C, Cadore JL, Montelaro RC (2004) Equine Infectious Anemia Virus (EIAV): what has HIV's country cousin got to tell us? Vet Res 35(4):485–512
Leroux C, Craigo JK, Issel CJ, Montelaro RC (2001) Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J Virol 75(10):4570–4583
Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, Berry CC, Ecker JR, Bushman FD (2006) Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog 2(6):e60
Lichtenstein DL, Craigo JK, Leroux C, Rushlow KE, Cook RF, Cook SJ, Issel CJ, Montelaro RC (1999) Effects of long terminal repeat sequence variation on equine infectious anemia virus replication in vitro and in vivo. Virology 263(2):408–417
Lim KH, Park ES, Kim DH, Cho KC, Kim KP, Park YK, Ahn SH, Park SH, Kim KH, Kim CW, Kang HS, Lee AR, Park S, Sim H, Won J, Seok K, You JS, Lee JH, Yi NJ, Lee KW, Suh KS, Seong BL, Kim KH (2018) Suppression of interferon-mediated anti-HBV response by single CpG methylation in the 5'-UTR of TRIM22. Gut 67(1):166–178
Ma J, Jiang C, Lin Y, Wang X, Zhao L, Xiang W, Shao Y, Shen R, Kong X, Zhou J (2009) In vivo evolution of the gp90 gene and consistently low plasma viral load during transient immune suppression demonstrate the safety of an attenuated equine infectious anemia virus (EIAV) vaccine. Arch Virol 154(5):867–873
Payne SL, La Celle K, Pei XF, Qi XM, Shao H, Steagall WK, Perry S, Fuller F (1999) Long terminal repeat sequences of equine infectious anaemia virus are a major determinant of cell tropism. J Gen Virol 80(Pt 3):755–759
Pion M, Jaramillo-Ruiz D, Martinez A, Munoz-Fernandez MA, Correa-Rocha R (2013) HIV infection of human regulatory T cells downregulates Foxp3 expression by increasing DNMT3b levels and DNA methylation in the FOXP3 gene. AIDS 27(13):2019–2029
Quinlivan M, Cook F, Kenna R, Callinan JJ, Cullinane A (2013) Genetic characterization by composite sequence analysis of a new pathogenic field strain of equine infectious anemia virus from the 2006 outbreak in Ireland. J Gen Virol 94(Pt 3):612–622
Robbins PB, Skelton DC, Yu XJ, Halene S, Leonard EH, Kohn DB (1998) Consistent, persistent expression from modified retroviral vectors in murine hematopoietic stem cells. Proc Natl Acad Sci USA 95(17):10182–10187
Saxonov S, Berg P, Brutlag DL (2006) A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA 103(5):1412–1417
Shan L, Yang HC, Rabi SA, Bravo HC, Shroff NS, Irizarry RA, Zhang H, Margolick JB, Siliciano JD, Siliciano RF (2011) Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J Virol 85(11):5384–5393
Wang X, Wang S, Lin Y, Jiang C, Ma J, Zhao L, Lv X, Wang F, Shen R, Zhou J (2011) Unique evolution characteristics of the envelope protein of EIAV(LN(4)(0)), a virulent strain of equine infectious anemia virus. Virus Genes 42(2):220–228
Wang X, Lin Y, Li Q, Liu Q, Zhao W, Du C, Chen J, Wang X, Zhou J (2016) Genetic evolution during the development of an attenuated EIAV vaccine. Retrovirology 13(1):9–9
Watt F, Molloy PL (1988) Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev 2(9):1136–1143
Wei L, Fan X, Lu X, Zhao L, Xiang W, Zhang X, Xue F, Shao Y, Shen R, Wang X (2009) Genetic variation in the long terminal repeat associated with the transition of Chinese equine infectious anemia virus from virulence to avirulence. Virus Genes 38(2):285–288
Wu X, Li Y, Crise B, Burgess SM (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300(5626):1749–1751
Zhong C, Hou Z, Huang J, Xie Q, Zhong Y (2015) Mutations and CpG islands among hepatitis B virus genotypes in Europe. BMC Bioinform 16:38
Acknowledgements
This study was supported by a grant from the Nanchong Vocational and Technical College fund for basic scientific research (Grant Nos. ZRA1904 and NZYBZ2002).
Author information
Authors and Affiliations
Contributions
YYY and WHY conducted the phylogenetic and sequence analyses. LQ conducted CpG analyses and wrote the manuscript. All authors have read and approved the fnal manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Takeshi Noda.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Q., Yu, YY. & Wang, HY. Comparative analysis of CpG islands in equine infectious anemia virus strains. Virus Genes 56, 339–346 (2020). https://doi.org/10.1007/s11262-020-01749-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-020-01749-1