Abstract
Background
The large family of pentatricopeptide repeat (PPR) proteins is widely distributed among land plants. Such proteins play vital roles in intron splicing, RNA editing, RNA processing, RNA stability and RNA translation. However, only a small number of PPR genes have been identified in rice.
Results
In this study, we raised a mutant from tissue-culture-derived plants of Oryza sativa subsp. japonica ‘Zhonghua 11’, which exhibited a lethal chlorosis phenotype from germination to the third-leaf stage. The mutant was designated seedling-lethal chlorosis 1 (slc1). The slc1 mutant leaves showed extremely low contents of photosynthetic pigments and abnormal chloroplast development, and were severely defective in photosynthesis. Map-based cloning of OsSLC1 revealed that a single base (G) deletion was detected in the first exon of Os06g0710800 in the slc1 mutant, which caused a premature stop codon. Knockout and complementation experiments further confirmed that OsSLC1 is responsible for the seedling-lethal chlorosis phenotype in the slc1 mutant. OsSLC1 was preferentially expressed in green leaves, and encoded a chloroplast-localized PPR protein harboring 12 PPR motifs. Loss-of-function of OsSLC1 affected the intron splicing of multiple group II introns, and especially precluded the intron splicing of rps16, and resulted in significant increase in the transcript levels of 3 chloroplast ribosomal RNAs and 16 chloroplast development-related and photosynthesis-related genes, and in significant reduction in the transcript levels of 1 chloroplast ribosomal RNAs and 2 chloroplast development-related and photosynthesis-related genes.
Conclusion
We characterized a novel chloroplast-localized PPR protein, OsSLC1, which plays a vital role in the intron splicing of multiple group II introns, especially the rps16 intron, and is essential for early chloroplast development and seedling survival in rice.
Similar content being viewed by others
Background
The chloroplast is a vital photosynthetic organelle for plant growth and development. The plant chloroplast is predicted to contain approximately 2500–3000 proteins, of which less than 10% are encoded by the chloroplast genome, the majority being encoded by the nuclear genome (Race et al. 1999; Armbruster et al. 2011). Introns are prevalent in plant chloroplast genomes, and intron splicing is an essential step prior to RNA translation.
The rice chloroplast genome is about 135 kb in size, consisting of approximately 128 genes, and 17 cis-spliced introns and one trans-spliced intron are distributed across 15 genes (Hiratsuka et al. 1989; Wu and Ge 2016). Among these introns, one in the trnL gene belongs to group I, with the remaining introns belonging to group II. However, the ability of these introns to perform self-splicing in vivo has been lost, and thus additional nuclear- and/or plastid-encoded splicing cofactors are required to accomplish intron splicing (Bonen and Vogel 2001; de Longevialle et al. 2010). Such cofactors include the plastid maturase matK, chloroplast RNA splicing and ribosome maturation proteins, pentatricopeptide repeat (PPR) proteins, RNA helicases, mitochondrial transcription termination factors, a RNase III domain-containing protein, WHAT IS THIS FACTOR 1 (WTF1) and a WHY and UMP kinase (de Longevialle et al. 2010; Quesada 2016; Nawaz and Kang 2017; Schmid et al. 2019). Of these cofactors, PPR proteins not only play vital roles in intron splicing, but also in RNA editing, RNA processing, RNA stability and RNA translation (Barkan and Small 2014).
A large family of PPR proteins is widely distributed among land plants. Approximately 477 PPR proteins are known in the rice nuclear genome (Lurin et al. 2004; Barkan and Small 2014). Generally, PPR proteins are divided into P (only containing PPR motifs) and PLS (containing PPR, PPR-like L (for long) and S (for short) motifs) subgroups, and on the basis of C-terminal motifs, the PLS subgroup is further subdivided into PLS, E/E+ (containing E/E+ motifs) and DYW (containing DYW motifs) subgroups (Lurin et al. 2004). To date, only a small number of PPR genes, comprising five restorer-of-fertility genes and approximately 25 additional PPR genes, have been identified in rice.
Among the identified PPR proteins, 11 are reported to be involved in intron splicing. OsOTP51 is involved in the intron splicing of atpF, especially ycf-2, in chloroplasts (Ye et al. 2012). The mitochondrion-localized OsMPR25 is involved in the intron splicing of atpF in chloroplasts (Toda et al. 2012; Yap et al. 2015). OsPPR4 (Asano et al. 2013) and OsWSL4 (Wang et al. 2017a) are both predominantly involved in the intron splicing of atpF, ndhA, rpl2 and rps12 in chloroplasts. The chloroplast-localized OsWSL is involved in the intron splicing of rpl2 in chloroplasts (Tan et al. 2014). The chloroplast-localized OsPPR6 is involved in the intron splicing of ycf3 in chloroplasts (Tang et al. 2017). The chloroplast-localized OsSLA4 is mainly involved in the intron splicing of atpF, ndhA, petB, rpl2, rpl16, rps12 and trnG in chloroplasts (Wang et al. 2018). The chloroplast-localized OsWSL5 is predominantly involved in the intron splicing of rpl2 and rps12 in chloroplasts (Liu et al. 2018). The chloroplast-localized OsPGL12 is mainly involved in the intron splicing of ndhA in chloroplasts (Chen et al. 2019). The mitochondrion-localized OsFLO10 is mainly involved in the trans-splicing of the nad1 intron 1 in mitochondria (Wu et al. 2019), and the nucleus-localized OsNPPR1 was reported to be involved in the intron splicing of a small number of nuclear-localized genes, many of which are mitochondrion-localized (Hao et al. 2019). The chloroplast-localized and/or mitochondrion-localized PPR proteins, such as OsPPR1 (Gothandam et al. 2005), OsOTP51, OsPPR4, OsWSL, OsALS3 (Lin et al. 2015a), OspTAC2 (Wang et al. 2016a), OsPPR6 (Tang et al. 2017), OsSLA4, OsWSL5 and OsPGL12, are essential for early chloroplast development in rice at the seedling stage.
In the present study, we characterized a novel PPR gene, OsSLC1, from a rice seedling-lethal chlorosis 1 (slc1) mutant. OsSLC1 encodes a chloroplast-localized P subgroup PPR protein harboring 12 PPR motifs. Analysis of loss-of-function of the slc1 mutant revealed that OsSLC1 plays a vital role in the intron splicing of multiple group II introns, especially the rps16 intron, and is essential for early chloroplast development and seedling survival in rice.
Results
The slc1 Mutant Exhibits a Seedling-Lethal Chlorosis Phenotype
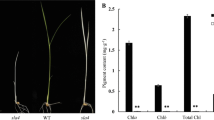
The slc1 mutant was obtained from tissue-culture-derived plants of rice Oryza sativa subsp. japonica ‘Zhonghua 11’, which exhibited a chlorosis phenotype from germination to the third-leaf stage, and seedlings gradually died off around 1 month after germination. The slc1 mutant at the second-leaf stage is shown in Fig. 1a. The chlorophyll a, chlorophyll b and carotenoid contents were extremely low in the slc1 mutant compared with the wild type (Fig. 1b).
Phenotypic characteristics of the slc1 mutant and measurement of leaf photosynthetic parameters in the wild type (WT) and slc1 plants at the seedling stage. a Phenotypes of the wild type and slc1 plants at the second-leaf stage. Scale bar, 2 cm. b Leaf photosynthetic pigment contents of the WT and slc1 plants at the third-leaf stage. Chl, chlorophyll; Car, carotenoid. c The net photosynthetic rate. d The stomatal conductance. e The intercellular CO2 concentration. f The transpiration rate. Three biological replicates were performed. Error bars are SDs. The asterisks indicate statistical significance between the wild type and slc1 mutant, as determined by Student’s t-test (** P < 0.01)
The slc1 Mutant Shows Severe Defects in Photosynthesis and Chloroplast Development
To investigate whether leaf photosynthesis was affected in the slc1 mutant, photosynthetic parameters were measured. The net photosynthetic rate, stomatal conductance and transpiration rate were significantly decreased (Fig. 1c, d, f), and the intercellular CO2 concentration was significantly increased in the slc1 mutant compared with the wild type (Fig. 2e). These results indicate that the slc1 mutant is severely defective in photosynthesis.
To investigate whether chloroplast development was affected in the slc1 mutant leaves, the chloroplast ultrastructure was observed. The leaf mesophyll cells contained large vesicles, and the chloroplasts exhibited normal shapes but accumulated some starch grains, and contained indistinct thylakoids and a small number of grana stacks in the slc1 mutant (Fig. 2b, d, f); in contrast, the leaf mesophyll cells contained normal-sized vesicles, and the chloroplasts contained distinct thylakoids and a large number of regular and dense grana stacks in the wild type (Fig. 2a, c, e). These results indicate that chloroplast development is severely impaired in the slc1 mutant.
Map-Based Cloning of OsSLC1
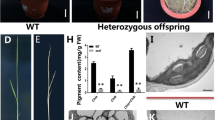
All heterozygous F1 plants showed a normal green phenotype and segregation occurred in F2 plants. The segregation ratio of 3:1 (green:chlorotic = 338:126; χ2 = 1.14 < χ20.05 = 3.84) indicates that a single recessive locus results in the seedling-lethal chlorosis phenotype in the slc1 mutant. The locus was designated OsSLC1.
Using 87 chlorotic mutants, the OsSLC1 locus was initially mapped to chromosome 6 between markers R5814 and R345 (Fig. 3a). To constrain the interval containing the OsSLC1 locus, an additional F2 population with 1244 chlorotic mutants was used for fine mapping. The OsSLC1 locus was mapped to a 50.7 kb interval between markers R3634–10 and R4329–4 on the BAC clone AP003634 (Fig. 3b). This region contains a total of nine putative open reading frames (Fig. 3c). Sequencing analysis revealed that a single-base (G) deletion was detected in the first exon of Os06g0710800 in the slc1 mutant (Fig. 3c), which caused a premature stop codon. Os06g0710800 was predicted to contain four exons with a coding sequence of 1479 bp. Accordingly, Os06g0710800 was selected as the candidate gene for OsSLC1.
Map-based cloning of OsSLC1. a The OsSLC1 locus was mapped to chromosome 6 (Chr. 6). b The OsSLC1 locus was fine-mapped to a 50.7 kb interval on the BAC clone AP003634. c Open reading frame of the candidate gene Os06g0710800 in the mapped region; the mutation site is indicated by a black upright triangle. d T0 mutant generated using the CRISPR/Cas9 system. e Independent T1 complementation plants. WT, wild type. Scale bars, 2 cm
To investigate whether the loss-of-function of Os06g0710800 results in the chlorosis phenotype in the slc1 mutant, the non-mutated Os06g0710800 gene of the wild type ‘Zhonghua 11’ was knocked out using a CRISPR/Cas9 system. Nine homologous T0 transgenic plants displayed the same seedling-lethal chlorosis phenotype. To further confirm the function of Os06g0710800, the OsSLC1-EGFP fusion cassette was transformed into the slc1 calli. A total of 19 of 21 T0 transgenic plants rescued the normal wild-type phenotype. The T0 knock-out line and independent T1 complementation plants are illustrated in Fig. 3d and e, respectively. These results indicate that Os06g0710800 is the candidate OsSLC1 gene, and is responsible for the seedling-lethal chlorosis phenotype in the slc1 mutant.
OsSLC1 Is Preferentially Expressed in Green Leaves and Encodes a Chloroplast-Localized Protein
To investigate the expression pattern of OsSLC1 in the wild type, quantitative real-time PCR was performed to analyze the relative expression level of OsSLC1 in a variety of tissues. OsSLC1 was predominantly expressed in the culm, sheath and green leaf, but was relatively highly expressed in green leaves (Fig. 4a).
Expression profile analysis of OsSLC1 in the wild type plant and subcellular localization of OsSLC1 in rice protoplasts. a Expression analysis of OsSLC1 in a variety of wild type tissues. RNAs were extracted from roots, culms, sheaths, leaves, young panicles (white) and green panicles at the heading stage. b Observation of free green fluorescent protein (GFP) signals in rice protoplasts. c Observation of OsSLC1-GFP fluorescent signals in rice protoplasts. Scale bars, 5 μm
The majority of PPR proteins contain a chloroplast- or mitochondrion-targeting peptide. To determine whether OsSLC1 was localized to the chloroplasts or mitochondria, the pOsSLC1-GFP vector was introduced into rice protoplasts. The green fluorescent signals from the OsSLC1-GFP fusion proteins overlapped with the chloroplast autofluorescences (Fig. 4b). These results indicate that OsSLC1 is localized to the chloroplasts.
OsSLC1 Is a Member of the P Subgroup of PPR Proteins
OsSLC1 was predicted to be a PPR protein composed of 492 amino acids. Protein sequence analysis showed that OsSLC1 contained a chloroplast transit peptide and 12 PPR motifs, and thus belonged to the P subgroup of the PPR family (Fig. 5a). Phylogenetic analysis showed that a class of unreported proteins, which were homologous to OsSLC1, were clustered into monocotyledon and dicotyledon groups (Fig. 5b). These results indicate that a novel class of unknown function of proteins homologous to OsSLC1 are widely distributed among angiosperms.
Structure and phylogenetic analysis of OsSLC1. a Predicted structure of OsSLC1. OsSLC1 contains a chloroplast transit peptide (cTP) and 12 PPR motifs (P). b Phylogenetic tree of OsSLC1 and its homologs. The protein sequences of A0A1D6CNW3 (Triticum aestivum) and A0A0E0Q2I6 (Oryza rufipogon) were downloaded from the UniProt database; all other sequences were downloaded from the National Center for Biotechnology Information database
The slc1 Mutant Is Defective in the Intron Splicing of Multiple Group II Introns, Especially the rps16 Intron
The P subgroup of PPR proteins are generally involved in intron splicing, RNA processing, RNA stability and RNA translation (Barkan and Small 2014; Manna 2015). Firstly, RT-PCR and quantitative real-time PCR were used to analyze the intron splicing of all 18 intron-containing chloroplast genes. The RT-PCR results showed that the intron splicing of rps16 was precluded, whereas other introns were naturally spliced in the slc1 mutant compared with the wild type (Fig. 6a). The quantitative real-time PCR results showed that the transcript levels of the spliced rps16 was severely reduced, and those of the spliced ndhA and petB were also variously reduced, whereas those of the spliced atpF, ndhB, petD, rpl2, rpl16, rps12, ycf3–1, ycf3–2, trnA, trnG, trnI, trnK and trnV were distinctly increased, and those of the other spliced genes analyzed did not change that much in the slc1 mutant compared with the wild type (Supplementary Figure S1). Further, quantitative real-time PCR was used to analyze the intron splicing efficiency of 18 intron-containing genes. The intron splicing efficiency of rps16 was extremely reduced, and those of atpF, ndhA, ndhB, petB, petD, rpl2, rps12, ycf3–1, ycf3–2, trnA, trnK and trnL were variously reduced (Fig. 6b), but most of their spliced mature transcripts were variously increased or not notably changed in the slc1 mutant compared with the wild type (Fig. 6a, Supplementary Figure S1). In addition, we examined 26 chloroplast RNA editing sites in the wild type and slc1 mutant. The majority of the editing sites were completely edited except for a few minor changes in the slc1 mutant compared with the wild type (Supplementary Table S1). Taken together, these results indicate that OsSLC1 plays a vital role in the intron splicing of multiple group II introns, especially the rps16 intron, rather than RNA editing, in rice.
Splicing analysis of chloroplast intron-containing transcripts in the wild type and slc1 mutant leaves at the third-leaf stage. a RT-PCR analysis of 18 intron-containing genes in the wild type and slc1 mutant leaves. U, Unspliced transcripts; S, Spliced transcripts. b Splicing efficiency analysis of chloroplast intron-containing transcripts in the wild type and slc1 mutant leaves. psbA was used as internal control, and relative transcript levels were analyzed using the 2-ΔΔCT method (Livak and Schmittgen 2001). Three biological replicates were performed. Values indicating the log2 ratios of spliced to unspliced transcripts in the slc1 mutant compared to the corresponding values in the wild type. Error bars are SDs. The asterisks indicate statistical significance between the wild type and slc1 mutant, as determined by Student’s t-test (* P < 0.05, ** P < 0.01)
Transcript Levels of Chloroplast Ribosomal RNAs, and Chloroplast Development-Related and Photosynthesis-Related Genes Are Altered in the slc1 Mutant
To investigate whether the expression levels of chloroplast and nuclear genes were affected in the slc1 mutant, chloroplast ribosomal RNAs and chloroplast development-related and photosynthesis-related genes were detected in the wild type and slc1 mutant. The transcript levels of 23S, 5S and 4.5S rRNAs were significantly increased, whereas those of 16S rRNA was significantly reduced in the slc1 mutant (Fig. 7a). The transcript levels of the plastid-encoded RNA polymerase (PEP) genes rpoA, rpoB, rpoC1 and rpoC2 were significantly increased, whereas those of the nuclear-encoded RNA polymerase gene RpoTp was hardly changed in the slc1 mutant (Fig. 7b). The transcript levels of the nuclear-encoded chloroplast large 50S and small 30S subunits genes RPL12, RPL13, RPL21, RPS6 and RPS20 were not notably changed, whereas those of the plastid-encoded large 50S and small 30S subunits genes rpl2, rpl16, rpl23, rps2, rps12 and rps15 were significantly increased in the slc1 mutant (Fig. 7b). The transcript levels of the chloroplast development-related and photosynthesis-related genes PORA and Cab2R were significantly reduced, whereas those of HEMD, CHLM, CAO1, petA, atpA and ndhB were significantly increased, and those of the other genes analyzed did not change that much in the slc1 mutant (Fig. 7c). These results indicate that OsSLC1 plays an important role in transcript expression of chloroplast development-related and photosynthesis-related genes in rice.
Quantitative real-time PCR analysis of genes associated with chloroplast development and photosynthesis in the wild type and slc1 mutant leaves at the third-leaf stage. a Transcript analysis of chloroplast 23S, 16S, 5S and 4.5S rRNAs. b Transcript analysis of plastid-encoded and nucleus-encoded chloroplast development-related genes. c Transcript analysis of chlorophyll synthesis-related and photosynthesis-related genes. OsActin1 and OsUbiquitin were used as double internal controls, and relative transcript levels were analyzed using the 2-ΔΔCT method (Livak and Schmittgen 2001). Three biological replicates were performed. Error bars are SDs. The asterisks indicate statistical significance between the wild type and slc1 mutant, as determined by Student’s t-test (** P < 0.01)
Discussion
PPR Proteins Are Essential for Normal Plant Growth and Development
Although the large PPR family is widely distributed among plants, only a small number of PPR proteins have been characterized. The majority of PPR proteins contain chloroplast- or mitochondrion-targeting peptides (Lurin et al. 2004), and some contain nucleus-targeting peptides (Ding et al. 2006; Hao et al. 2019). PPR proteins commonly participate in RNA metabolism in organelles or the nucleus, and are essential for normal plant growth and development.
Mutations of PPR genes in rice typically cause a variety of abnormal phenotypes, such as seedling lethality, abnormal seedling growth, delayed seed germination, retarded growth, dwarfism, defective seed development, embryo lethality and sterility. For example, mutations of chloroplast-localized PPR genes generally cause seedling-lethal albino or chlorosis phenotypes under natural conditions, as is the case for OsPPR1 (Gothandam et al. 2005), OsOTP51 (Ye et al. 2012), OsPPR4 (Asano et al. 2013), OsASL3 (Lin et al. 2015a), OspTAC2 (Wang et al. 2016a), OsPPR6 (Tang et al. 2017) and OsSLA4 (Wang et al. 2018). Loss-of-function of OsSLC1 also causes a seedling-lethal chlorosis phenotype in the slc1 mutant. Mutations of mitochondrion-localized PPR genes typically cause retarded growth, dwarfism, defective seed development, as is the case for OsOGR1 (Kim et al. 2009), OsEMP5 (Liu et al. 2013), OsSMK1 (Li et al. 2014) and OsPPS1 (Xiao et al. 2018).
OsSLC1 Is Essential for Early Chloroplast Development and Seedling Survival
The chloroplast possesses its own protein translation system. The bacterial-type chloroplast 70S ribosome, consisting of 30S small and 50S large subunits, is essential for protein translation in chloroplasts (Harris et al. 1994). The 30S ribosomal subunit, composed of 24 proteins, recruits the 16S rRNA to initiate the translation of mRNA, whereas the large 50S ribosomal subunit, composed of 33 proteins, recruits the 23S, 5S and 4.5S rRNAs to activate the initiation of translation of mRNA (Yamaguchi et al. 2000; Yamaguchi and Subramanian 2000; Tiller and Bock 2014; Bieri et al. 2017). The 16S rRNA plays a vital role in mRNA binding and stabilization of codon-anticodon interaction (Bieri et al. 2017). Previous studies indicate that loss-of-function of genes that encode the 30S small or 50S large subunits always causes abnormal chloroplast development and seedling growth in rice.
Mutation of OsASL1, which encodes the 30S small subunit protein RPS20, causes impaired chloroplast development and a seedling-lethal albino phenotype (Gong et al. 2013). Mutations of OsWLP1 (Song et al. 2014) and OsTCD11 (Wang et al. 2017b), which encode the 50S large subunit protein RPL13 and the 30S small subunit protein RPS6, respectively, causes impaired chloroplast development and an albino seedling phenotype at low temperature. Mutations of OsASL2 (Lin et al. 2015b) and OsAL1 (Zhao et al. 2016), which encode the 50S large subunit protein RPL21 and RPL12, respectively, cause impaired chloroplast development and seedling-lethal albino phenotype. Mutation of OsWGL2, which encodes the 30S small subunit protein RPS9, causes impaired chloroplast development and an albino seedling phenotype (mutants generated by CRISPR/Cas9) (Qiu et al. 2018).
Loss-of-function of OsSLC1 especially precluded the intron splicing of rps16 in the slc1 mutant (Fig. 6a, b), which indicates that the post-transcriptional processing and translation of rps16 was blocked in the mutant. Given the deficiency in Rps16 protein, the 30S small subunit is unable to recruit 16S rRNAs, which thus could be degraded, and result in severe reduction in the transcript levels of 16S rRNAs in the slc1 mutant (Fig. 7a). Therefore, we infer that chloroplasts in the slc1 mutant failed to assemble the normal 70S ribosomes, which resulted in severe defects in protein synthesis. As a result of the impairment in protein synthesis, the transcript levels of PEP genes, such as rpoA, rpoB, rpoC1 and rpoC2, and those of plastid-encoded ribosome genes, such as rpl2, rpl16, rpl23, rps2, rps12 and rps15, accumulated significantly in the slc1 mutant (Fig. 7b). In addition, given the impediment in protein synthesis, chloroplasts in the slc1 mutant possessed abnormal thylakoids and few grana stacks (Fig. 2d, f), which resulted in few photosynthetic pigments bound by abnormal thylakoids in the slc1 mutant (Fig. 1b). Thus, photosynthesis was severely impaired in the slc1 mutant (Fig. 2). In addition, the transcript levels of the photosynthesis-related genes PORA and Cab2R were significantly reduced in the slc1 mutant (Fig. 7c). Taken together, the present results reveal that OsSLC1 is indispensable for early chloroplast development and normal seedling survival in rice.
Multiple Factors May Be Involved in the Intron Splicing of rps16
In the previous studies, several intron splicing factors have been identified to be involved in the intron splicing of rps16, such as ZmCRS2, ZmCAF1, ZmWTF1 and OsCFM3 (Asakura et al. 2008; Jenkins et al. 1997; Kroeger et al. 2009; Ostheimer et al. 2003). Such factors are also found in large ribonucleoprotein complexes containing other factors. In maize, CRS2 was found in a large ribonucleoprotein complex containing CAF1 (Ostheimer et al. 2003), and WTF1 in a large ribonucleoprotein complex containing both CAF1 and CFM3 (Kroeger et al. 2009). Thus, we infer that OsSLC1, OsCAF1, OsCFM3, OsCRS2 and OsWTF1 together with other factors may form a ribonucleoprotein complex to perform the intron splicing of rps16 in rice chloroplast. However, further work is needed to test this hypothesis.
Conclusion
We described a novel chloroplast-localized P subgroup PPR protein, OsSLC1, which harbors 12 PPR motifs. OsSLC1 functions in the intron splicing of multiple chloroplast group II introns, especially the rps16 intron, and plays a vital role in early chloroplast development and seedling survival in rice.
Methods
Plant Materials and Growth Conditions
The slc1 mutant was obtained from tissue-culture-derived plants of rice Oryza sativa subsp. japonica ‘Zhonghua 11’. The heterozygous slc1 plants were preserved to reproduce the slc1 mutants. All rice plants used in this study were grown in fields at Chongqing or in growth chambers under 12 h light (10,000 lx) at 30 °C and 12 h dark at 26 °C.
Photosynthetic Pigment and Photosynthetic Parameter Measurements
Leaf photosynthetic pigment contents of the wild type and slc1 mutant at the third-leaf stage were measured as previously described (Lichtenthaler 1987; Wang et al. 2016b). Fresh leaves (approximately 0.1 g fresh weight) at the third-leaf stage were cut into small pieces and soaked in 25 mL of 95% ethanol for 24 h at room temperature in the dark. Pigment contents were measured with a UV-1800PC (Mapada) spectrophotometer at 663, 645 and 470 nm. Three biological replicates were performed. Microsoft Office Excel 2016 was used to analyze data, and Student’s two-tailed paired t-test was used to determine the statistical significance of data between the wild type and slc1 mutant, the same as below. Leaf photosynthetic parameters, consisting of net photosynthetic rate, stomatal conductance, intercellular CO2 concentration and transpiration rate, were measured at the third-leaf stage from 09:00 to 11:00 with an LI-6400 portable photosynthesis system (LI-COR, Lincoln, NE, USA) in accordance with the manufacturer’s instructions. Three biological replicates were performed.
Transmission Electron Microscopy
The chloroplast ultrastructure of the wild type and slc1 mutant leaves at the third-leaf stage was examined as previously described (Liu et al. 2007). Leaf samples were fixed with 2.5% glutaraldehyde and 1% OsO4, dehydrated in an ethanol series, and finally embedded in Spurr resin. The fixed and embedded samples were stained with uranyl acetate and alkaline lead citrate and then observed with a H-7500 transmission electron microscope (Hitachi, Tokyo, Japan).
Map-Based Cloning of OsSLC1
The heterozygous slc1 plants were crossed with Oryza sativa subsp. indica ‘Jinhui 1’ for genetic analysis. The F1 seeds were harvested from each individual plant, and mutants segregating in the F2 population were selected for gene mapping. Sequence polymorphisms between Oryza sativa subsp. japonica ‘Nipponbare’ and subsp. indica ‘93–11’ were used to develop insertion/deletion and simple sequence repeat molecular markers (Shen et al. 2004). Gene annotation and primer design for DNA and cDNA sequencing were performed on the basis of information obtained from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) database and Gramene (http://gramene.org/) databases. Multiple sequence alignment was performed with Vector NTI Advance 10 (Invitrogen, USA; http://www.invitrogen.com/). All primers used in this study are listed in Supplementary Table S2.
Vector Construction for the CRISPR/Cas9 System and Genetic Complementation
A modified tRNA-processing strategy based on the CRISPR/Cas9 system was used for knockout of OsSLC1 in the wild type. The procedures were followed as previously described (Wang et al. 2018). Two gRNA target sites (CCGTGGGAGTCCTACGACCGCGG, CGGTGTCAAGCCGAATACCCCGG) were designed to construct the OsSLC1-Cas9 vector, which was transformed into the wild-type (‘Zhonghua 11’) calli using an Agrobacterium-mediated method (Hiei and Komari 2008).
For complementation of the slc1 mutant, the full-length coding sequences of OsSLC1 and EGFP were cloned into the pCAMRubi2 vector digested with EcoRI and HindIII to generate the OsSLC1-EGFP vector, which was driven by the OsUbiquitin2 promoter (Wang et al. 2000; Wang et al. 2016b). The OsSLC1-EGFP vector was transformed into the slc1 calli using an Agrobacterium-mediated method (Hiei and Komari 2008).
Sequence and Phylogenetic Analysis
Subcellular location prediction was performed using TargetP (http://www.cbs.dtu.dk/services/TargetP/) and ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). Protein structure prediction was performed using NCBI, PROSITE (http://prosite.expasy.org/prosite.html), and Smart (http://smart.embl-heidelberg.de/). Proteins homologous to OsSLC1 were obtained by BLAST and downloaded from NCBI and UniProt (http://www.uniprot.org/). A neighbor-joining tree was constructed with the software MEGA v7.0, and statistical support for the tree topology was assessed by means of a bootstrap analysis with 1000 replicates (Kumar et al. 2016).
RNA Preparation and Quantitative Real-Time PCR Analysis
Total rice RNAs were extracted from roots, culms, sheaths, leaves, young panicles (white) and green panicles of the wild type plants at the heading stage using an RNAprep Pure Plant Kit (Tiangen Biotech). First-strand cDNA was synthesized from 1 μg total RNA using a PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time, TaKaRa). Quantitative real-time PCR was conducted using a TB Green Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa) on a Bio-Rad CFX96 system according to the manufacturer’s instructions. The genes used for quantitative real-time PCR were consistent with the previous studies (Wang et al. 2016b; Zhang et al. 2017; Wang et al. 2018). OsActin1 and OsUbiquitin were used as double internal controls, and relative transcript levels were analyzed using the 2–ΔΔCT method (Livak and Schmittgen 2001). Three biological replicates were performed.
Subcellular Localization of OsSLC1
The full-length coding sequence of OsSLC1 was fused to the N-terminus of GFP in the pAN580 vector digested with SpeI and BamHI to generate the pOsSLC1-GFP vector. The pOsSLC1-GFP vector and the empty vector pAN580 were transformed into rice protoplasts as previously described (Zhang et al. 2011). A confocal laser scanning microscope (LSM700, Zeiss, Jena, Germany) was used to observe GFP fluorescence.
Chloroplast RNA Editing and RNA Splicing Analysis
Total RNA was extracted from the wild type and slc1 mutant leaves at the third-leaf stage using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China), and treated with Recombinant DNase I (RNase-free) (TaKaRa, Tokyo, Japan). First-strand cDNA was synthesized with random hexamers using a PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa) (for RT-PCR analysis), and synthesized with RT primer mix using a PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time, TaKaRa) (for quantitative real-time PCR analysis). cDNA was diluted to 50 ng·μL− 1 and used for PCR amplification with PrimeSTAR Max DNA Polymerase (TaKaRa). A total of 18 intron splicing analysis and 26 C-to-U RNA editing analysis were performed as previously described (Corneille et al. 2000; Inada et al. 2004; Tan et al. 2014; Zhang et al. 2017; Wang et al. 2018). The RT-PCR primers used for intron splicing and RNA editing analysis were consistent with the previous study (Wang et al. 2018). The quantitative real-time PCR primers used for intron splicing and unsplicing analysis were shown in the Supplementary Table S2. Each pair of primers were designed to cross each intron for splicing analysis, whereas at least one primer was designed to locate each intron for unsplicing analysis. Quantitative real-time PCR was performed as mentioned above. psbA was used as internal control, and relative transcript levels were analyzed using the 2–ΔΔCT method (Livak and Schmittgen 2001). Three biological replicates were performed. The intron splicing efficiencies were analyzed using the log2 ratios of spliced to unspliced transcripts in the slc1 mutant compared to the corresponding values in the wild type.
Abbreviations
- PPR:
-
pentatricopeptide repeat
- CRISPR/Cas9:
-
clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9
- PEP:
-
plastid-encoded RNA polymerase
References
Armbruster U, Pesaresi P, Pribil M, Hertle A, Leister D (2011) Update on chloroplast research: new tools, new topics, and new trends. Mol Plant 4:1–16
Asakura Y, Bayraktar OA, Barkan A (2008) Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA 14:2319–2332
Asano T, Miyao A, Hirochika H, Kikuchi S, K-i K (2013) A pentatricopeptide repeat gene of rice is required for splicing of chloroplast transcripts and RNA editing of ndhA. Plant Biotech 30:57–64
Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65:415–442
Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N (2017) The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J 36:475–486
Bonen L, Vogel J (2001) The ins and outs of group II introns. Trends Genet 17:322–331
Chen L, Huang L, Dai L, Gao Y, Zou W, Lu X, Wang C, Zhang G, Ren D, Hu J, Shen L, Dong G, Gao Z, Chen G, Xue D, Guo L, Xing Y, Qian Q, Zhu L, Zeng D (2019) PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in rice. Plant Cell Physiol 60:587–598
Corneille S, Lutz K, Maliga P (2000) Conservation of RNA editing between rice and maize plastids: are most editing events dispensable? Mol Gen Genet 264:419–424
de Longevialle AF, Small ID, Lurin C (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3:691–705
Ding YH, Liu NY, Tang ZS, Liu J, Yang WC (2006) Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18:815–830
Gong X, Jiang Q, Xu J, Zhang J, Teng S, Lin D, Dong Y (2013) Disruption of the rice plastid ribosomal protein S20 leads to chloroplast developmental defects and seedling lethality. G3-Genes Genom Genet 3:1769–1777
Gothandam KM, Kim E-S, Cho H, Chung Y-Y (2005) OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 58:421–433
Hao Y, Wang Y, Wu M, Zhu X, Teng X, Sun Y, Zhu J, Zhang Y, Jing R, Lei J (2019) The nuclear-localized PPR protein OsNPPR1 is important for mitochondrial function and endosperm development in rice. J Exp Bot. https://doi.org/10.1093/jxb/erz226
Harris EH, Boynton JE, Gillham NW (1994) Chloroplast ribosomes and protein synthesis. Microbiol Rev 58:700–754
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3:824–834
Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY et al (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217:185–194
Inada M, Sasaki T, Yukawa M, Tsudzuki T, Sugiura M (2004) A systematic search for RNA editing sites in pea chloroplasts: an editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol 45:1615–1622
Jenkins BD, Kulhanek DJ, Barkan A (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9:283–296
Kim SR, Yang JI, Moon S, Ryu CH, An K, Kim KM, Yim J, An G (2009) Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J 59:738–749
Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A (2009) A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Nati Acad Sci 106:4537–4542
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li XJ, Zhang YF, Hou M, Sun F, Shen Y, Xiu ZH, Wang X, Chen ZL, Sun SSM, Small I, Tan BC (2014) Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J 79:797–809
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Method Enzymol 148:350–382
Lin D, Gong X, Jiang Q, Zheng K, Zhou H, Xu J, Teng S, Dong Y (2015a) The rice ALS3 encoding a novel pentatricopeptide repeat protein is required for chloroplast development and seedling growth. Rice 8:17
Lin D, Jiang Q, Zheng K, Chen S, Zhou H, Gong X, Xu J, Teng S, Dong Y (2015b) Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol 17:599–607
Liu WZ, Fu YP, Hu GC, Si HM, Zhu L, Wu C, Sun ZX (2007) Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 226:785–795
Liu X, Lan J, Huang Y, Cao P, Zhou C, Ren Y, He N, Liu S, Tian Y, Nguyen T (2018) WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J Exp Bot 69:3949–3961
Liu YJ, Xiu ZH, Meeley R, Tan BC (2013) Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing in maize. Plant Cell 25:868–883
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16:2089–2103
Manna S (2015) An overview of pentatricopeptide repeat proteins and their applications. Biochim 113:93–99
Nawaz G, Kang H (2017) Chloroplast- or mitochondria-targeted DEAD-box RNA helicases play essential roles in organellar RNA metabolism and abiotic stress responses. Front Plant Sci 8:871
Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A (2003) Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J 22:3919–3929
Qiu Z, Chen D, He L, Zhang S, Yang Z, Zhang Y, Wang Z, Ren D, Qian Q, Guo L, Zhu L (2018) The rice white green leaf 2 gene causes defects in chloroplast development and affects the plastid ribosomal protein S9. Rice 11:39
Quesada V (2016) The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol Plant 157:389–399
Race HL, Herrmann RG, Martin W (1999) Why have organelles retained genomes? Trends Genet 15:364–370
Schmid L-M, Ohler L, Möhlmann T, Brachmann A, Muiño JM, Leister D, Meurer J, Manavski N (2019) PUMPKIN, the sole plastid UMP kinase, associates with group II introns and alters their metabolism. Plant Physiol 179:248–264
Shen YJ, Jiang H, Jin JP, Zhang ZB, Xi B, He YY, Wang G, Wang C, Qian LL, Li X, Yu QB, Liu HJ, Chen DH, Gao JH, Huang H, Shi TL, Yang ZN (2004) Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol 135:1198–1205
Song J, Wei X, Shao G, Sheng Z, Chen D, Liu C, Jiao G, Xie L, Tang S, Hu P (2014) The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol Biol 84:301–314
Tan JJ, Tan ZH, Wu FQ, Sheng PK, Heng YQ, Wang XH, Ren YL, Wang JL, Guo XP, Zhang X, Cheng ZJ, Jiang L, Liu XM, Wang HY, Wan JM (2014) A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol Plant 7:1329–1349
Tang J, Zhang W, Wen K, Chen G, Sun J, Tian Y, Tang W, Yu J, An H, Wu T (2017) OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol Biol 95:345–357
Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7:1105–1120
Toda T, Fujii S, Noguchi K, Kazama T, Toriyama K (2012) Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. Plant J 72:450–460
Wang D, Liu H, Zhai G, Wang L, Shao J, Tao Y (2016a) OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. J Genet Genomics 43:601–608
Wang JL, Jiang JD, Oard JH (2000) Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.). Plant Sci 156:201–211
Wang WJ, Zheng KL, Gong XD, Xu JL, Huang JR, Lin DZ, Dong YJ (2017b) The rice TCD11 encoding plastid ribosomal protein S6 is essential for chloroplast development at low temperature. Plant Sci 259:1–11
Wang Y, Ren Y, Zhou K, Liu L, Wang J, Xu Y, Zhang H, Zhang L, Feng Z, Wang L, Ma W, Wang Y, Guo X, Zhang X, Lei C, Cheng Z, Wan J (2017a) WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front Plant Sci 8:1116
Wang ZW, Lv J, Xie SZ, Zhang Y, Qiu ZN, Chen P, Cui YT, Niu YF, Hu SK, Jiang HZ, Ge SZ, Trinh H, Lei KR, Bai WQ, Zhang Y, Guo LB, Ren DY (2018) OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul 84:249–260
Wang ZW, Zhang TQ, Xing YD, Zeng XQ, Wang L, Liu ZX, Shi JQ, Zhu XY, Ma L, Li YF, Ling YH, Sang XC, He GH (2016b) YGL9, encoding the putative chloroplast signal recognition particle 43 kDa protein in rice, is involved in chioroplast development. J Integr Agr 15:944–953
Wu M, Ren Y, Cai M, Wang Y, Zhu S, Zhu J, Hao Y, Teng X, Zhu X, Jing R (2019) Rice FLOURY ENDOSPERM 10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytol 223:736–750
Wu Z, Ge S (2016) The whole chloroplast genome of wild rice (Oryza australiensis). Mitochondrial DNA Part A 27:1062–1063
Xiao H, Zhang Q, Qin X, Xu Y, Ni C, Huang J, Zhu L, Zhong F, Liu W, Yao G, Zhu Y, Hu J (2018) Rice PPS1 encodes a DYW motif-containing pentatricopeptide repeat protein required for five consecutive RNA-editing sites of nad3 in mitochondria. New Phytol 220:878–892
Yamaguchi K, Kv K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 30S subunit of an organelle ribosome (chloroplast). J Biol Chem 275:28455–28465
Yamaguchi K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 50S subunit of an organelle ribosome (chloroplast). J Biol Chem 275:28466–28482
Yap A, Kindgren P, Colas des Francs-Small C, Kazama T, Tanz SK, Toriyama K, Small I (2015) AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J 81:661–669
Ye JW, Gong ZY, Chen CG, Mi HL, Chen GY (2012) A mutation of OSOTP51 leads to impairment of photosystem I complex assembly and serious photo-damage in rice. J Integr Plant Biol 54:87–98
Zhang Y, Su JB, Duan S, Ao Y, Dai JR, Liu J, Wang P, Li YG, Liu B, Feng DR, Wang JF, Wang HB (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30
Zhang ZG, Cui XA, Wang YW, Wu JX, Gu XF, Lu TG (2017) The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol Plant 10:86–98
Zhao DS, Zhang CQ, Li QF, Yang QQ, Gu MH, Liu QQ (2016) A residue substitution in the plastid ribosomal protein L12/AL1 produces defective plastid ribosome and causes early seedling lethality in rice. Plant Mol Biol 91:161–177
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (31701390, 31370349), Project on Basic Research and Frontier Exploration of Chongqing (cstc2018jcyjAX0234), Special Project on Performance Incentive Guidance of Chongqing Scientific Research Institution (cstc2018jxjl80021), Fundamental Research Funds of Chongqing (2016cstc-jbky-00524), and Foundation of Discovery and Utilization of Green, High-quality and High-efficiency Rice Germplasm Resources (NKY-2018QC03), the Agricultural Science and Technology Innovation Program, Shenzhen Science and Technology Program (2017050414212249).
Author information
Authors and Affiliations
Contributions
JL, LS and YC designed and performed the experiments, and analyzed the data. YH, XY, SX, WB, MH, HW, KL, YY, SG and HT performed the experiments and analyzed the data. YZ, LG and ZW designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no potential competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1 : Figure S1
. Quantitative real-time PCR analysis of chloroplast spliced genes in the wild type and slc1 mutant leaves at the third-leaf stage. Table S1. Analysis of chloroplast RNA editing (C to U) in the wild type and slc1 mutant. Table S2. Primer sequences used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lv, J., Shang, L., Chen, Y. et al. OsSLC1 Encodes a Pentatricopeptide Repeat Protein Essential for Early Chloroplast Development and Seedling Survival. Rice 13, 25 (2020). https://doi.org/10.1186/s12284-020-00385-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-020-00385-5