Abstract
Viral infections are commonly associated with honey bee (Apis mellifera) colony mortality. Using metagenomics, we previously identified 8 viruses from populations of honey bees and 11 other bee species around the world. These viruses had not been previously been described as bee-infecting viruses, and belong to viral families that are not commonly described in bees. To provide a fine-scale characterization of these viruses in the USA, we screened bees from the 2015 USDA National Honey Bee Disease Survey. Two viruses are widespread, and thus likely require further characterization, while four may represent emerging or under surveyed infections. We also compare different approaches for screening samples for viral infections. This study demonstrates the importance of creating and maintaining large-scale collections for the broader research community.
Similar content being viewed by others
1 Introduction
Large-scale declines have been documented in multiple insect pollinator populations, which raises significant concerns, as pollinators are critical for agricultural production and maintaining healthy ecosystems (LeBuhn et al. 2012; Potts et al. 2010). Many factors contribute to these declines, including pathogens, parasites, pesticide exposure, altered landscapes contributing to inadequate nutrition, and human-driven global climate change, as well as their synergistic effects (Goulson et al. 2015). Viral pathogens are a particularly interesting driver of bee declines, as pathogens have been found to cause symptoms ranging from asymptomatic to lethal (Grozinger and Flenniken 2019), synergize with many other stressors (Di Prisco et al. 2013; Doublet et al. 2015), and readily infect numerous bee species, and thus are transmitted across co-foraging pollinator communities (Tehel et al. 2016). Recent studies have used metagenomic approaches to characterize populations of viruses infecting different bee species, and have identified several “novel” viruses that have previously not been identified in bees (Galbraith et al. 2018; Remnant et al. 2017; Schoonvaere et al. 2018).
One recent metagenomic survey, Galbraith et al. (2018), studied the viromes of bee communities across nine countries spanning five continents, identifying 127 sequences representing previously undescribed viruses in bees. Many corresponded to (+)ssRNA viruses, which are most commonly found in honey bees, and were assigned names based on their similarities to known viral families (Table I). These (+)ssRNA viruses included the Dicistro-like, Noda-like, Seco-like, and Tymo-like viruses. The Galbraith et al. (2018) study also identified other RNA virus types rarely described in bees previously, including (−)ssRNA (bee rhabdovirus) and dsRNA (Partiti-like virus). Additionally, the authors provided the first evidence of circular ssDNA viruses (denoted Circo-like viruses 1 and 2) infecting bees. Furthermore, many of these potential viruses were identified in both Apis mellifera honey bees and co-foraging, non-Apis bee specimens, suggesting the potential for a wide distribution of these viruses across bee communities (Table I).
Since the viruses from Galbraith et al. (2018) were identified in bee populations across the five continents assessed, it suggests that these pathogens are broadly distributed and potentially quite prevalent. However, since metagenomics studies often examine only a small number of samples, it is difficult to extrapolate from these studies the prevalence and distribution of these viruses on an ecologically relevant scale. For example, while some viruses were identified within the USA, this survey was not an exhaustive screen, only testing in three of 50 states (Galbraith et al. 2018). A much more detailed sampling effort is required to fully assess virus distribution and the potential impact on pollinator populations.
To track disease associated with losses in managed honey bee colonies within the USA, the US Department of Agriculture (USDA) Animal and Plant Health Inspection Service (APHIS) National Honey Bee Disease Survey (NHBS) was initiated in 2009, and collections and screenings continue annually (Traynor et al. 2016). Samples include bees collected from hundreds of apiaries in locations across the USA, including approximately 25–40 states and the territory of Puerto Rico. Live bee samples are sent to the USDA–Agricultural Research Service (ARS) Beltsville Bee Research Laboratory, where molecular techniques are employed to test for the presence of seven common known bee viruses (Gisder and Genersch 2017). The archived samples from APHIS NHBS serve as an important collection that can be used to monitor the presence of newly identified viruses as well as uncovering the possible historical presence of viruses thought to be recently introduced into the US bee populations. Indeed, the NHBS collection was recently used to document the rapid spread of a second master variant of deformed wing virus (DWV), known as DWV-B or Varroa destructor virus-1, across the USA in only 6 years (Ryabov et al. 2017).
As the Galbraith et al. (2018) study found that the highest number of novel viruses were identified in North America, we utilized the collections from the APHIS NHBS in 2015 to screen for the eight recently identified viruses, providing finer scale information of their prevalence and distribution within the USA.
2 Materials and methods
Samples
At the time of this study, the NHBS from 2015 collected the highest number of samples compared to other years to that date, and was therefore selected for this study; conveniently, this was also the same year as the bee collections for Galbraith et al. (2018). Sample collection is described in Traynor et al. (2016). Briefly, each sample consisted of pools of 50 bees from a single apiary, for a total of 800 samples from 35 states and Puerto Rico. RNA extraction (Qiagen RNEasy, Hilden, Germany) and cDNA synthesis with 2 μg total RNA (U Superscript II reverse transcriptase, Invitrogen, Carlsbad, California, USA) were previously conducted as in Traynor et al. (2016). Prior to our study, cDNA samples were kept at – 20 °C for long-term storage.
Sample pooling
To expedite screening, cDNA from samples collected within the same state were pooled together, with 800 original cDNA samples combined into a total of 68 pools. The pooling strategy consisted of adding 2 μl of original cDNA to each pool, with 10–15 apiaries per pool, and 1–2 pools per state. However, 3 pools were created for California, as this state had a total of 43 apiaries due to the high density of apiaries in the state used for pollination services. Sample ID per pool can be found in the supplemental tables (see Supplementary Table 1 “Samples with Pooling ID”).
Viral detection
Primers used in this study can be found in Table II. These primers were designed to be unique to the replicase regions of the novel putative viruses and were made using Primer3 and confirmed with Primer Blast. These primers were previously utilized in Galbraith et al. (2018), with 7/8 (all but Circo-like 1) confirmed to amplify the correct target sequence via Sanger sequencing at the Genomics Core Facility at Penn State University.
Virus presence was assessed via quantitative PCR (qPCR). The following conditions were used for the qPCR assessment: each well-contained 5 μl PowerUp Syber Green Master mix (Applied Biosystems, Foster City, California, USA), 1 μl each of 10 μM Forward and Reverse Primer, 1 μl H2O, and 2 μl cDNA (approximately 53 ng) for a total of 10 μl per reaction. qPCR was conducted using a 7900HT Fast Real-Time PCR system (Applied Biosystems) under the following parameters: 50°C for 2 min, 95°C for 10 min, then cycle 40 × 95°C for 15 s and 59 °C for 1 min, and then a dissociation stage step for melting curve analysis. When possible, positive controls were included on each plate, consisting of the original cDNA from Galbraith et al. (2018). Negative controls on each plate consisted of a no-cDNA H2O sample. For 7/8 virus targets, R2 efficiency was calculated from 1:10 serial dilutions of positive controls or samples—all R2 values were greater than 0.95 (Supplementary Table “Pool ct values”).
Ct value was used to score the presence or absence of virus within a sample by our defined detection threshold. Detection threshold for virus identification was determined by the lowest measured Ct in an H2O (negative control) sample, corresponding to a Ct of 30.71. A sample with a Ct below 30.71 was considered a “Positive” identification for the novel virus, and above as “Negative” (for Ct values for positive and negative controls, see Supplementary Tables). Additionally, melting curve temperatures were measured and Sanger sequencing was used for confirmation of target amplification and exclude samples with possible off-target amplification due to primer–dimers or other artifacts. In the case of Circo-like 2 and Tymo-like virus, melting temperatures were difficult to distinguish between samples and negative controls. While it is not standard protocol to report on data that similarly amplified in the negative control, we chose to report the potential presence of these viruses, as positive controls were sequence confirmed (and had much lower Ct values, 21 and 10), and detection in NHBS samples was low, 1 or 5 locations, respectively. Nevertheless, these results should be interpreted with caution.
3 Results
The novel viruses in this study showed extreme differences in frequency, ranging from no detection to positive identification in nearly every location tested (Table III, Figure 1). Two viruses were the most common: bee rhabdovirus (BRV) and Partiti-like virus (PLV) were identified in 33/36 and 28/36 locations, respectively. Tymo-like virus (TLV) was detected in five states, and Seco-like virus (SLV) was detected in two. The two Circo-like viruses (CLV1 and CLV2) were detected in one state each. Two viruses, Noda-like virus (NLV) and Dicistro-like virus (DLV), were not positively identified in any state. Ct values can be found in Table III and Supplemental Table “Pool Ct values”.
Average infection levels of newly identified viruses across the USA. Bold lines indicate average, with top and bottom of boxes indicating the first and third median values. Bars indicate SE, and outliers indicated with circles. The horizontal line indicates the presence threshold, 30.71, determined by the negative control. Individual Ct values can be found in Supplementary Table “Pool Ct values”. Plots were generated using the ggplot function in RStudio (R Core Team 2017).
To identify whether the pooling strategy distorted signal from low occurrence viruses, the original, un-pooled cDNA was assessed for all viruses except for BRV and PLV, which were already easily identified across the USA, and any additional screen at the apiary level would likely demonstrate a highly similar dispersal. These apiary level experiments revealed fewer locations with positive identifications (Table IV, Supplementary Table “Apiary Ct values”), and identified locations other than those from the pooled sample experiments. In this screen, TLV and SLV were detected in two separate apiaries in Washington. CLV1 was not positively identified in any un-pooled sample. CLV2 was initially detected in pooled cDNA from Florida but was identified here in un-pooled cDNA from Kentucky and Washington samples. NLV continued to be undetected in un-pooled samples, likely indicative of little to no presence in US apiaries. DLV, on the other hand, was positively identified in Washington, despite no detection in samples pooled by state.
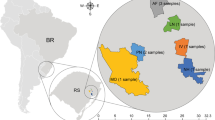
The viruses themselves have different ranges across locations in the pooled NHBS samples (Figure 2). All states assessed and Puerto Rico have had at least one of the eight viruses identified within their state, with most states (n = 21) having two viruses in their state. Florida, with the most positive identifications in the pooled samples, had four viruses positively identified, while Washington was the highest for the un-pooled samples, also having four viruses detected.
Maps of virus presence across the US reveal distribution differences of the eight newly identified viruses. Status of each state is indicated by color, with virus “Present,” “Absent,” or untested. A virus was deemed “Present” if at least one pooled cDNA sampled met the detection threshold of lower than 30.71. Plots were generated using the “fiftystater” package in ggplot in RStudio (Murphy 2019; R Core Team 2017), and edited in Adobe Photoshop to include Puerto Rico.
4 Discussion and conclusion
In this study, we detected the presence of six of eight newly described bee viruses in honey bee apiaries across the USA (Figure 2). In particular, two viruses (BRV and PLV) are broadly distributed, and thus further tests should be conducted to evaluate the impact of infections with these viruses on bees. Four other viruses (TLV, SLV, CLV1, and CLV2) were detected in only a few locations, and thus may represent emerging or transient infections. Additionally, we demonstrated varying distributions of viruses across the states, with some states exhibiting substantially more viral diversity than others. Overall, these results provide further validation of using metagenomics approaches, such as Galbraith et al. 2018, to characterize bee viral populations but also demonstrate the critical need for the types of finer scale mapping studies conducted in this study. Indeed, some of the most prevalent viruses in the metagenomics study were not prevalent in the NHBS samples (e.g., SLV), and vice versa (e.g., BRV). Importantly, this study demonstrates the value of creating and maintaining large-scale collections of bee population samples that are broadly accessible to the research community.
Our results provide critical insights for designing large-scale virus screens in bee populations. First, for low prevalence viruses, screening individual apiaries provided similar limited ability to detect viruses as screening samples pooled from 10 to 15 apiaries. In other words, screening for low-level viruses in samples of 50 bees provided similar information as screening 500–750 bees at a time. Thus, if detailed spatial information is not necessary, pooling provides an economical and efficient method for tracking viral frequency across larger regions and many more samples. In some cases, it is possible that viral RNA in one apiary was diluted by pooling with uninfected apiaries. This may explain why some states, like Oregon in the case of BRV, can score as “negative” for virus but be surrounded by states identified as “positive.” Furthermore, while our threshold approach was efficient, it was also conservative (as some may consider any amplification whatsoever as indicative of the presence of a virus) and may have resulted in false negatives for locations with low levels of viruses.
Secondly, we were able to detect circular ssDNA viruses (CLV1 and CLV2) in this study. Since Traynor et al. (2016) extracted RNA from these samples, and included a DNaseI digestion to remove genomic DNA, detection of ssDNA viruses from isolated RNA is likely indicative of transcription of these viral genomes. This provides evidence that these putative viruses are indeed infecting honey bees and actively transcribing. Thus, while Galbraith et al. (2018) used specialized approaches to obtain both RNA and DNA viruses from their samples, for a simple screening using smaller sequence fragments, using extracted RNA may be sufficient. While these results indicate active infection for the DNA viruses, it would be valuable to conduct additional analyses of virus replication of the other viruses in the study.
BRV and PLV can be found across the USA. In the case of BRV, this result may come with little surprise, as it has been identified globally and in multiple species. Its first published identification came in 2016 in populations of honey bees and Varroa destructor mites from Israel, where it was identified as “Farmington virus” (Levin et al. 2016). Soon after, rhabdoviruses were identified in honey bees and bumble bees in additional locations worldwide, further demonstrating global distribution and cross-species prevalence, and inspiring a rename to “Apis” rhabdovirus then to “Bee” rhabdovirus (Levin et al. 2017; Remnant et al. 2017). Subsequently, BRV and other putative rhabdoviruses in bees have been described in numerous additional studies (Galbraith et al. 2018; Regan et al. 2018; Schoonvaere et al. 2018; Thaduri et al. 2018). Despite identification in bee communities worldwide, no work has yet described the impact of BRV on bee health and decline, nor any co-infection dynamics with other widespread bee viruses, nor the potential role of Varroa destructor as a viral vector.
PLV is an interesting case, as viruses in the Partitiviridae family most often target plants and fungi (Nibert et al. 2014), although recent characterizations of invertebrate RNA viromes have described viruses in the Partitiviridae family in other arthropod viromes (Flynn and Moreau 2019; Garigliany et al. 2017; Shi et al. 2016). Not only is this virus found in bees but is distributed widely across apiaries in the USA. The presence of plant-targeting viruses in pollinators may not be unexpected, as other plant viruses have been detected in honey bees (Li et al. 2014b; although see Miller et al. 2014 and Li et al. 2014a), providing an effective plant virus surveillance method by using high throughput sequencing in honey bees (Roberts et al. 2018). Additional studies are needed to determine whether PLV in bees indicates that bees are serving as vector of a plant pathogen (Whitfield et al. 2015), PLV is a generalist pathogen infecting both kingdoms (Gray and Banerjee 1999), or PLV represents an evolutionary host-switching event from plants to bees (Longdon et al. 2014). Why PLV and BRV are so common yet only characterized by metagenomic sequencing, rather than after observation of symptomatic infection, may be indicative of a mild, asymptomatic infection phenotype; alternatively, severely infected bees may perish from infection before any potential disease is observed. Still, infection dynamics and evolutionary history of these viruses remain undescribed, and their ubiquity merits future characterization.
Viruses in the Tymoviridae and Secoviridae families often target plants, similar to the Partitiviridae family of viruses (Thompson et al. 2014), although TLV and SLV were not nearly as prevalent as PLV in this study. TLV was the third most common virus identified in the pooled samples in this study, but was still only detected in five locations. However, these locations ranged from the South to the Northwest US, indicating a potentially large distribution but low prevalence of the virus. SLV was one of the most prevalent viruses identified in the global metagenomic study (Galbraith et al. 2018), but was rarely found in this study.
The last two novel putative viruses from the Galbraith et al. (2018) study, NLV and DLV, were not very common in the Galbraith et al. (2018) samples, and not identified in the pooled NHBS samples, though DLV was detected in one un-pooled sample. Both of these viral families are known to infect insects (Odegard et al. 2010)—members of the Dicistroviridae family include common viruses targeting bees, such as black queen cell virus and Israeli acute paralysis virus (Bonning 2009). Since infection dynamics of these putative viruses is unknown, it is unclear if the viruses are rare due to a severe infection phenotype, leading to rare pockets of outbreaks, or represent a new pathogen to managed bees, with the potential to spread across the nation or globe (McMahon et al. 2018). Future years of the NHBS can be used to monitor and track the spread of all of these and other viruses.
Different states exhibited clear differences in viral diversity. In the case of the pooled samples, four of the six detected viruses were identified in Florida, whereas most states only had 1–2 detectable viruses. The cause of this is not immediately clear, as all statewide pools included a similar number of apiaries (10 to 15, and each Florida pool covered 11 apiaries), but Southern states might have better overwintering survival, year-round brood production, and additional ectoparasites like the small hive beetle, which may allow for viruses to persist in the environment (Doke et al. 2015; Eyer et al. 2009). Washington state, conversely, had the most detected viruses in the un-pooled samples, and this state differs from Florida dramatically in its climatic conditions. It is possible that factors such as stressful winters and high densities of crops utilizing migratory honey bee pollination services may increase pathogen levels and transmission. Future studies to characterize ecological factors influencing the epidemiology of these novel viruses will provide insight into the explanation behind the prevalence and distribution of these viruses across the USA and globe.
This study demonstrates the tremendous value of large-scale, long-term, accessible collections which can be used to screen for newly identified or emerging pathogens and parasites in honey bees or other insects, and furthermore assess their role in insect population declines (Leather 2018; Sánchez-bayo and Wyckhuys 2019). Our results provide insight into the approaches which can be used to efficiently screen these collections, namely the advantages and disadvantages of pooling samples and the ability to identify both RNA and DNA viruses using a single screen. Importantly, this study has demonstrated that two viruses in particular, BRV and PLV, are widely distributed in US bee populations, but belong to viral families grossly understudied in bees. Future studies should evaluate the transmission and infection dynamics of these viruses within both managed and wild bee populations.
References
Bonning B.C. (2009) The dicistroviridae: An emerging family of invertebrate viruses. Virol. Sin. 24(5), 415–427. https://doi.org/10.1007/s12250-009-3044-1
Di Prisco G., Cavaliere V., Annoscia D., Varricchio P., Caprio E., Nazzi F. et al (2013) Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. 110(46), 18466–18471. https://doi.org/10.1073/pnas.1314923110
Doke M.A., Frazier M., Grozinger C.M. (2015) Overwintering honey bees: biology and management. Curr. Opin. Insect Sci. 10, 185–193
Doublet V., Labarussias M., de Miranda J.R., Moritz R.F.A., Paxton R.J. (2015) Bees under stress: Sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ. Microbiol. 17(4), 969–983. https://doi.org/10.1111/1462-2920.12426
Eyer M., Chen Y.P., Schafer M.O., Pettis J., Neumann P. (2009) Small hive beetle, Aethina tumida, as a potential biological vector of honeybee viruses. 40, 419–428
Flynn P.J., Moreau C.S. (2019) Assessing the Diversity of Endogenous Viruses Throughout Ant Genomes. 10(May). https://doi.org/10.3389/fmicb.2019.01139
Galbraith D.A., Fuller Z.L., Ray A.M., Brockmann A., Frazier M., Gikungu M.W. et al (2018) Investigating the viral ecology of global bee communities with high-throughput metagenomics. Sci. Rep. (June), 1–14. https://doi.org/10.1038/s41598-018-27164-z
Garigliany M., Taminiau B., Cadar D., Gilliaux G., Hue M., Desmecht D. et al (2017) Moku Virus in Invasive Asian Hornets, Belgium, 2016. 23(12), 10–13
Gisder S., Genersch E. (2017) Viruses of commercialized insect pollinators. J. Invertebr. Pathol. 147, 51–59. https://doi.org/10.1016/j.jip.2016.07.010
Goulson D., Nicholls E., Botías C., Rotheray E.L. (2015) Bee declines driven by combined Stress from parasites, pesticides, and lack of flowers. Science 347(6229). https://doi.org/10.1126/science.1255957
Gray S.M., Banerjee N. (1999) Mechanisms of Arthropod Transmission of Plant and Animal Viruses. 63(1), 128–148
Grozinger, C. M., & Flenniken, M. L. (2019). Bee Viruses: Ecology , Pathogenicity , and Impacts.
Leather S.R. (2018) “ Ecological Armageddon ” – more evidence for the drastic decline in insect numbers. 172, 1–3. https://doi.org/10.1111/aab.12410
LeBuhn G., Droege S.A.M., Connor E.F., Gemmill-herren B., Potts S.G., Minckley R.L. et al (2012) Detecting Insect Pollinator Declines on Regional and Global Scales. 27(1), 113–120. https://doi.org/10.1111/j.1523-1739.2012.01962.x
Levin S., Galbraith D., Sela N., Erez T., Grozinger C.M., Chejanovsky N. (2017) Presence of Apis rhabdovirus-1 in populations of pollinators and their parasites from two continents. Front. Microbiol. 8(DEC), 1–11. https://doi.org/10.3389/fmicb.2017.02482
Levin S., Sela N., Chejanovsky N. (2016) Two novel viruses associated with the Apis mellifera pathogenic mite Varroa destructor. Sci. Rep. 6, 1–9. https://doi.org/10.1038/srep37710
Li J.L., Cornman R.S., Evans J.D., Pettis J.S., Zhao Y., Murphy C. et al (2014a) Reply to “ Conclusive Evidence of Replication of a Plant Virus in Honeybees Is Lacking ,”. 5(3), 3–5. https://doi.org/10.1128/mBio.01250-14.Copyright
Li J.L., Cornman R.S., Evans J.D., Pettis J.S., Zhao Y., Murphy C. et al (2014b) Systemic Spread and Propagation of a Plant-Pathogenic Virus in European Honeybees , Apis mellifera. 5(1), 1–11. https://doi.org/10.1128/mBio.00898-13.Editor
Longdon B., Brockhurst M.A., Russell C.A., Welch J.J., Jiggins F.M. (2014) The Evolution and Genetics of Virus Host Shifts. 10(11). https://doi.org/10.1371/journal.ppat.1004395
McMahon D.P., Wilfert L., Paxton R.J., Brown M.J.F. (2018) Emerging Viruses in Bees: From Molecules to Ecology. Advances in Virus Research, 1st edn. Elsevier Inc. https://doi.org/10.1016/bs.aivir.2018.02.008
Miller W.A., Dolezal A.G., Toth A.L. (2014) Conclusive Evidence of Replication of a Plant Virus in Honeybees Is Lacking Conclusive Evidence of Replication of a Plant Virus in Honeybees Is. https://doi.org/10.1128/mBio.00985-14.Rights
Murphy, W. (2019). fiftystater: Map Data to Visualize the Fifty U.S. States with Alaska and Hawaii Insets. R package version 1.0.1. https://github.com/wmurphyrd/fiftystater
Nibert M.L., Ghabrial S.A., Maiss E., Lesker T., Vainio E.J., Jiang D., Suzuki N. (2014) Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 188, 128–141. https://doi.org/10.1016/j.virusres.2014.04.007
Odegard A., Banerjee M., Johnson J.E. (2010) Flock House Virus: A Model System for Understanding Non-Enveloped Virus Entry and Membrane Penetration. In Johnson J.E. (Ed.), Cell Entry by Non-Enveloped Viruses (pp. 1–22). Springer Berlin Heidelberg, Berlin. https://doi.org/10.1007/82_2010_35
Potts S.G., Biesmeijer J.C., Kremen C., Neumann P., Schweiger O., Kunin W.E. (2010) Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 25(6), 345–353. https://doi.org/10.1016/j.tree.2010.01.007
R Core Team (2017) R: A language and environment forstatistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Regan T., Barnett M.W., Laetsch D.R., Bush S.J., Wragg D., Budge G.E. et al (2018) Characterisation of the British honey bee metagenome. Nat. Commun. 9(1), 4995. https://doi.org/10.1038/s41467-018-07426-0
Remnant E.J., Shi M., Buchmann G., Blacquière T., Holmes E.C., Beekman M., Ashe A. (2017) A Diverse Range of Novel RNA Viruses in Geographically Distinct Honey Bee Populations Emily. 91(16), 1–19
Roberts J.M.K., Ireland K.B., Tay W.T., Paini D. (2018) Honey bee-assisted surveillance for early plant virus detection. Ann. Appl. Biol. 173(3), 285–293. https://doi.org/10.1111/aab.12461
Ryabov E.V., Childers A.K., Chen Y., Madella S., Nessa A., VanEngelsdorp D., Evans J.D. (2017) Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci. Rep. 7(1), 1–10. https://doi.org/10.1038/s41598-017-17802-3
Sánchez-bayo F., Wyckhuys K.A.G. (2019) Worldwide decline of the entomofauna: A review of its drivers. 232(January), 8–27. https://doi.org/10.1016/j.biocon.2019.01.020
Schoonvaere K., Smagghe G., Francis F., de Graaf D.C. (2018) Study of the metatranscriptome of eight social and solitary wild bee species reveals novel viruses and bee parasites. Front. Microbiol. 9(FEB), 1–12. https://doi.org/10.3389/fmicb.2018.00177
Shi M., Lin X., Tian J., Chen L., Chen X., Li C. et al (2016) Article Redefining the invertebrate RNA virosphere. Nat. Publ. Group 540(7634), 539–543. https://doi.org/10.1038/nature20167
Tehel A., Brown M.J.F., Paxton R.J. (2016) Impact of managed honey bee viruses on wild bees. Curr. Opin. Virol. 19, 16–22. https://doi.org/10.1016/j.coviro.2016.06.006
Thaduri S., Locke B., Granberg F., de Miranda J.R. (2018) Temporal changes in the viromes of Swedish varroa-resistant and varroa-susceptible honeybee populations. PLoS ONE, in press, 1–17. https://doi.org/10.1371/journal.pone.0206938
Thompson J.R., Kamath N., Perry K.L. (2014) An Evolutionary Analysis of the Secoviridae Family of Viruses. 9(9). https://doi.org/10.1371/journal.pone.0106305
Traynor K.S., Rennich K., Forsgren E., Rose R., Pettis J., Kunkel G. et al (2016) Multiyear survey targeting disease incidence in US honey bees. Apidologie 47(3), 325–347. https://doi.org/10.1007/s13592-016-0431-0
Whitfield A.E., Falk B.W., Rotenberg D. (2015) Insect vector-mediated transmission of plant viruses. 480, 278–289. https://doi.org/10.1016/j.virol.2015.03.026
Acknowledgments
We would like to thank members of the Grozinger lab for critical reading of the manuscript and Damie Pak for assistance with figures.
Funding
This project was funded by a grant from the US Department of Agriculture Animal and Plant Health Inspection Service (#15-8130-0501 and #16-8130-0501) awarded to CMG. A. Ray was supported by funding from the Molecular, Cellular, and Integrative Biosciences Intercollege Graduate Training Program in the Huck Institutes of the Life Sciences at Penn State University.
Author information
Authors and Affiliations
Contributions
AR, JFIM, DG, CR, and CG conceived research and designed experiments; DL, RR, DE, and JE performed previous sample collection; AR performed experiments; All authors read and approved the manuscript.
Corresponding author
Additional information
Manuscript editor: Michelle L. Flenniken
Répartition des virus récemment identifiés infectant les abeilles dans les populations d'abeilles ( Apis mellifera ) gérées aux États-Unis.
Apis mellifera / enquête sur la maladie / virus / USA.
Verbreitung von kürzlich identifizierten Bienenviren in imkerlich gehaltenen Honigbienenpopulationen ( Apis mellifera ) in den USA.
Apis mellifera / Krankheitsstudie / Viren / USA
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1.
(XLSX 148 kb)
Rights and permissions
About this article
Cite this article
RAY, A.M., LOPEZ, D.L., ITURRALDE MARTINEZ, J.F. et al. Distribution of recently identified bee-infecting viruses in managed honey bee (Apis mellifera) populations in the USA. Apidologie 51, 736–745 (2020). https://doi.org/10.1007/s13592-020-00757-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-020-00757-2