Abstract
Prionospio Malmgren 1867 and Aurospio Maciolek 1981 (Annelida: Spionidae) are polychaete genera commonly found in the deep sea. Both genera belong to the Prionospio complex, whose members are known to have limited distinguishing characters. Morphological identification of specimens from the deep sea is challenging, as fragmentation and other damages are common during sampling. These issues impede investigations into the distribution patterns of these genera in the deep sea. In this study, we employ two molecular markers (16S rRNA and 18S) to study the diversity and the distribution patterns of Prionospio and Aurospio from the tropical North Atlantic, the Puerto Rico Trench and the central Pacific. Based on different molecular analyses (Automated Barcode Gap Discovery, GMYC, pairwise genetic distances, phylogenetics, haplotype networks), we were able to identify and differentiate 21 lineages (three lineages composed solely of GenBank entries) that represent putative species. Seven of these lineages exhibited pan-oceanic distributions (occurring in the Atlantic as well as the Pacific) in some cases even sharing identical 16S rRNA haplotypes in both oceans. Even the lineages found to be restricted to one of the oceans were distributed over large regional scales as for example across the Mid-Atlantic Ridge from the Caribbean to the eastern Atlantic (> 3389 km). Our results suggest that members of Prionospio and Aurospio may have the potential to disperse across large geographic distances, largely unaffected by topographic barriers and possibly even between oceans. Their high dispersal capacities are probably explained by their free-swimming long-lived planktonic larvae.
Similar content being viewed by others
Introduction
The genus Prionospio Malmgren 1867 is one of the most diverse and speciose taxon within the Spionidae (Guggolz et al. 2018; Paterson et al. 2016). Prionospio is morphologically not well defined, even after several revisions and the erection of closely related subgenera and new genera in a Prionospio complex (Foster 1971; Maciolek 1985; Sigvaldadottir 1998; Wilson 1990). Currently, the Prionospio complex comprises the genera Aurospio Maciolek 1981, Laubieriellus Maciolek 1981a, Orthoprionospio Blake and Kudenov 1978, Prionospio Malmgren 1867, Streblospio Webster 1879, and Paraprionospio Caullery 1914. Most of these revisions were mainly based on shallow-water species, but Prionospio can also be regarded a typical deep-sea genus, often found in high abundances (Blake et al. 2018; Guggolz et al. 2018; Guggolz and Meißner pers. observations).
Despite the typical occurrence of Prionospio in deep-sea samples, the number of reported species is rather limited (Glover and Fauchald 2018: 29 species—http://www.marinespecies.org/deepsea/). Only a few generic characters are available to distinguish between the genera of the Prionospio complex; often, only the arrangement of the branchiae is important for the characterization of different subgenera and genera (Paterson et al. 2016). These characters seem to be sufficient to identify specimens in appropriate conditions, but the morphological identification of these soft-bodied annelids from deep-sea samples is often difficult. Due to their fragility, the majority of specimens from these depths are incomplete or damaged (Bogantes et al. 2018; Guggolz et al. 2018, 2019). For example, the genus Aurospio is mainly distinguished from Prionospio by the number of the branchiae and on which segment they are beginning (Sigvaldadóttir and Mackie 1993), but these appendages are often lost or damaged during sampling procedures.
Species of both genera, Prionospio and Aurospio, are reported to be widespread (Paterson et al. 2016) or even cosmopolitan (Mincks et al. 2009) (e.g., Aurospio dibranchiata Maciolek 1981). A wide dispersal potential in the abyssal has been reported for many benthic invertebrates (Etter et al. 2011; Linse and Schwabe 2018; Schüller and Ebbe 2007); however, these distribution patterns, based solely on morphological taxonomic identification, have to be treated with caution. Recent studies, employing molecular tools, often indicated a more complex scenario. Several of these presumably widespread species were found to be composed of a number geographically restricted and morphologically cryptic species (Bickford et al. 2007; Vrijenhoek 2009) or simply misidentified (Álvarez-Campos et al. 2017; Nygren et al. 2018; Sun et al. 2016). Hence, hypothesizing distribution patterns in the deep sea is still challenging, and integrative approaches, which combine morphological and molecular techniques, are essential to identify and delimit species (Glover et al. 2016; Hutchings and Kupriyanova 2018). One important aspect of the present study is to examine the diversity and the dispersal capacity of Prionospio and Aurospio in the Vema-Fracture-Zone (VFZ). Both genera, Prionospio and Aurospio, are supposed to have planktonic larvae and thus a potential for a widespread geographic distribution (Wilson 1991; Young 2003). We investigate the dispersal along the VFZ and test for barrier effect of the Mid-Atlantic Ridge (MAR) as this underwater mountain ridge is often postulated to represent a topographic barrier for distribution of benthic invertebrates (Bober et al. 2018; McClain et al. 2009; Priede et al. 2013). However, a barrier effect of the MAR on the spionid Laonice Malmgren 1867 has recently been rejected (Guggolz et al. 2019). The herein studied Prionospio and Aurospio will be another important step towards understanding the distribution patterns of species in the deep sea along the MAR. In addition, a potential pan-oceanic distribution is analyzed, by comparing DNA sequences of specimens from the VFZ (tropical Atlantic) with those of the Clarion-Clipperton Fracture Zone (CCZ) from the central Pacific.
Material and methods
Collection and identification of specimens
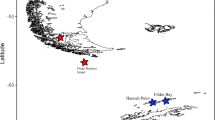
During the VEMA-Transit expedition in December 2014 to January 2015, 197 of the 332 analyzed specimens were collected from the tropical North Atlantic and the Puerto Rico Trench (Fig. 1: VFZ and PRT). Detailed information about sample treatment and sampling localities are described in Guggolz et al. (2018) and Devey (2015). Four areas were defined for samples from the Atlantic according to the geographical position as following: the eastern part of the Vema-Fracture Zone (eVFZ), extending eastwards from the MAR in the Cape Verde Basin; the western part of the Vema Fracture Zone (wVFZ), extending westwards from the MAR in the Demerara Basin; the Vema Transform Fault (VTF), located between these two areas in the MAR; the Puerto Rico Trench (PRT), located in the shallower part of the trench near Puerto Rico. Maximum distances within areas varied between 276 km (wVFZ) and 1298 km (eVFZ). The eastern-most and western-most studied sites were separated by 4610 km.
The Clarion-Clipperton Fracture Zone (CCZ) is a vast area (about 6 million km2) in the Equatorial Pacific Ocean with high commercial interest because of the presence of polymetallic nodules in the seabed between 4000 and 5000 m depth. The International Seabed Authority (ISA) is in charge of management of deep-sea mineral resources and of protection of marine environment in areas beyond national jurisdiction (Lodge et al. 2014). The ISA provides licenses to the contractors that intend to explore mineral deposits in, e.g., the CCZ. To get and keep an exploration contract for an area, the contractor is required to carry out surveys and fauna inventories (Lodge et al. 2014). Furthermore, the ISA administrates the regional environmental management plan across the CCZ, so-called Areas of Particular Environmental Interest (APEI). Sequences of 122 specimens morphologically assigned to Prionospio and Aurospio from eight exploration contract areas and one APEI were included (Fig. 1: the German exploration contract area ‘BGR’, Russia and Poland among other countries ‘IOM’, Belgium ‘GSR’, French ‘Ifremer’ & ‘Ifremer-2’, Singapore ‘OMS’, Britain UK-1 and the APEI-6). Of these, 53 specimens were collected on the two United Kingdom Seabed Resources Ltd. (UKSR) cruises AB01 and AB02 to the UK-1 exploration contract area stratum A and stratum B, the OMS contract area, and the APEI-6. Details on sampling methods are given in Glover et al. (2016). Maps and metadata from UK-1 stratum A have been published in earlier taxonomical work on macrofaunal material from this cruise (Dahlgren et al. 2016; A. Glover et al. 2016; Wiklund et al. submitted, 2017). In addition, 69 specimens morphologically assigned to Prionospio and Aurospio from BGR, IOM, GSR, and Ifremer were sampled using box-corers (0.25 m2) or epibenthic sledge during EcoResponse SO239 cruise on board of the RV Sonne in March/April 2015 funded by JPI Oceans framework (Martínez Arbizu and Haeckel 2015).
All specimens were sorted and identified at least to genus level (Prionospio or Aurospio) using dissecting and compound microscopes. Specimens have been deposited in the collection of the Center of Natural History (Universität Hamburg, Germany), Ifremer (France), and Natural History Museum London (Supplement 1).
The map of the sampling areas (Fig. 1) was created using ArcGIS 10.4.1 (www.esri.com).
DNA extraction, PCR amplification, sequencing, and alignment
For the VFZ specimens, one or two parapodia were dissected and transferred into 30 μl of 10% Chelex 100 solution in purified water, incubated for 30 min at 56 °C and 10 min at 99 °C. Polymerase chain reactions (PCR) were performed with a total volume of 15 μl consisting of 1.5 μl DNA extract, 7.5 μl AccuStart II PCR ToughMix (Quanta Bio, Germany), 0.6 μl of each primer (10 mmol), 0.3 μl of GelTrack loading dye (QuantaBio, Germany), and 4.8 μl Millipore H2O. Fragments of mitochondrial (16S) and nuclear (18S) rRNA genes were amplified (see Table 1 for list of primers) with initial denaturation step of 94 °C for 3 min, followed by 35 cycles of 30 s at 94 °C, 45 s at 43 °C, and 45 s at 72 °C, followed by a final elongation step for 5 min at 72 °C. Success of amplification was determined via gel electrophoresis on 1% agarose/TAE gel. For sequencing, 8 μl of the PCR products was purified using FastAP (1.6 μl; 1 U/μl) and Exonuclease I (0.8 μl; 20 U/μl) (Thermo Fisher Scientific, Germany) with an incubation time of 37 °C for 15 min followed by 15 min at 85 °C. For some of the specimens from the CCZ (BGR, IOM, GSR and Ifremer), DNA extractions were realized with NucleoSpin Tissue (Macherey-Nagel) kit and PCR amplifications as following into 25-μl mixtures, including 5 μl of Green GoTaq® Flexi Buffer (final concentration of 1×), 2.5 μl of MgCl2 solution (final concentration of 2.5 mM), 0.5 μl of PCR nucleotide mix (final concentration of 0.2 mM each dNTP), 9.875 μl of nuclease-free water, 2.5 μl of each primer (final concentration of 1 μM), 2 μl template DNA, and 0.125 U of GoTaq® G2 Flexi DNA Polymerase (Promega). The temperature profile was 95 °C/240 s, 94 °C/30 s, 52 °C/60 s, 72 °C/75 s *35 cycles, 72 °C/480 s, 4 °C. Purified PCR products were sent to Macrogen Europe, Inc. (Amsterdam-Zuidoost, Netherlands) for sequencing. The remaining specimens from the CCZ (APEI-6, OMS, UK-1) were extracted with DNeasy Blood and Tissue Kit (Qiagen) using a Hamilton Microlab STAR Robotic Workstation. PCR mixtures contained 1 μl of each primer (10 μM), 2 μl template DNA, and 21 μl of Red Taq DNA Polymerase 1.1× MasterMix (VWR) in a mixture of total 25 μl. The PCR amplification profile consisted of initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 94 °C for 45 s, annealing at 55 °C for 45 s, extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. PCR products were purified using Millipore Multiscreen 96-well PCR Purification System, and sequencing was performed on an ABI 3730XL DNA Analyser (Applied Biosystems) at Natural History Museum Sequencing Facility, using the same primers as in the PCR reactions.
In total, 331 specimens were successfully sequenced for 16S rRNA and 63 specimens for 18S. Additionally, the mitochondrial COI gene and the nuclear 28S and ITS genes were tested with considerably worse success and therefore deliberately omitted. 18S is known to show some degree of divergence, even at species level in spionids, and thus successfully used for supporting mitochondrial markers (Guggolz et al. 2019; Meißner et al. 2014; Meißner and Blank 2009). Sequences were assembled and corrected with Geneious 6.1.8 (Kearse et al. 2012; http://www.geneious.com) and deposited in GenBank (for accession numbers, see Supplement Table 1). The obtained sequences of the different gene fragments were aligned separately using MAFFT (Katoh and Standley 2013; V 7.402) implemented with CIPRES Science Gateway V.3.3 (Miller et al. 2010; www.phylo.org).
Initial identification of species, phylogenetic analyses, and haplotype networks
To obtain a first estimation of the number of species present in our data set, the Automated Barcode Gap Discovery (Puillandre et al. 2012; ABGD) was conducted with 16S rRNA. The ABGD identifies potential barcoding gaps separating hypothetical species, which is based on the assumption that interspecific genetic distances are larger than intraspecific distances. The ABGD analysis was run on the web-based version of the software (http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html). Pairwise uncorrected p-distances were used for the analyses (Table 2), which were calculated with MEGA7 (Kumar et al. 2016), including all available sequences for each gene, respectively. Standard settings for ABGD were kept, except for Pmin (0.005), the numbers of steps (100), and the relative gap width (X = 0.5). In the following, we will use the term lineages rather than species for the units delimited by ABGD, as not all of these might correspond to actual species. Additionally, the single locus tree-based species delimitation analysis GMYC (general mixed Yule coalescent; Pons et al. 2006) was used to support the results of the ABGD. For the GMYC analyses, an ultrametric tree was calculated with BEAST 2.4.6 (Bouckaert et al. 2014) employing the YULE coalescence model for 50*106 generations, retaining every 5000th tree. In this analysis, each haplotype was included only once. Convergence was assessed with Tracer v1.6 and TreeAnnotator v2.4.6 was used to obtain the final tree. GMYC was run in R, version 3.6.1 (R Core Team 2018) using standard settings for the single threshold analysis.
To assess the phylogenetic relationships among the studied specimens and to assess whether the lineages suggested by ABGD are monophyletic, phylogenetic analyses were performed with Bayesian inference and maximum likelihood. Both gene fragments were analyzed separately, as well as concatenated with MrBayes (Ronquist and Huelsenbeck 2003; version 3.2). The maximum likelihood was performed using Randomized Axelerated Maximum Likelihood (Stamatakis 2014; RAxML v.8.2.10) on XSEDE with rapid bootstrapping (1000 iterations) via the CIPRES Science Gateway V.3.3 (Miller et al. 2010; www.phylo.org). Whether a given phylogenetic hypothesis can be rejected by the data is often not obvious from the tree topology, especially in cases where some branches are not significantly supported. We used the approximately unbiased test (Shimodaira 2002) as implemented in the program CONSEL (Shimodaira and Hasegawa 2001) to test the statistical significance of likelihood differences between the best ML topology and the ML topology, in which the genus Aurospio was constrained to be monophyletic. The constrained ML tree and the site-wise likelihoods associated with each topology (necessary for conducting the approximately unbiased test) were calculated with GARLI 2.0 (Zwickl 2006). Due to missing sequences of the 18S genes for a large part of the analyzed specimens and the very few mutations, only the phylogenetic analysis of the 16S rRNA gene is included. For the 16S rRNA analysis, different spionids that are not assigned to the Prionospio complex were chosen as outgroups (Supplement 1). Additionally, available 16S rRNA sequences of Prionospio and Aurospio from GenBank were included in the analyses (Supplement 1). Four chains were run for 5*107 generations, with sampling every 1200th generation, and discarding the first 25% as burn-in. Thus, the convergence chain runs were validated using TRACER v.1.7.1 (Rambaut et al. 2018). The GTR + I + G substitution model was identified by MEGA7 as the best fitting model under the AIC criterion.
To better visualize the geographic distribution of the genetic diversity, median-joining haplotype networks were generated with Network 5.0.0.3 (http://fluxus-engineering.com/) and popART 1.7 (Bandelt et al. 1999) for each gene fragment. In 16S rRNA, networks were calculated separately for each lineage. The generated haplotype networks were redrawn with Adobe Illustrator CS6.
Analyses of population differentiation were performed with Arlequin 3.5 (Excoffier and Lischer 2010) for lineages with sufficiently large specimen numbers (at least four specimens per site). Pairwise Φst was calculated different lineages identified as Aurospio cf. dibranchiata, Aurospio sp. S, and Prionospio sp. B, sp.H (16S rRNA). For Aurospio cf. dibranchiata, the areas eVFZ, wVFZ, and CCZ, for Aurospio sp. S the areas eVFZ and CCZ and for Prionospio sp. B the areas eVFZ, VTF and CCZ and for Prionospio sp. G the sites 2 and 4 were compared (Tables 3 and 4).
Results
Alignment
The alignment of the 16S rRNA fragment included a total of 331 sequences with a minimum length of 442 base pairs (bp), of which 237 bp were variable and 223 bp were parsimony informative. The alignment of the 18S fragment consisted of 59 sequences with 845 bp minimum length, of which 25 bp were variable and 20 bp were parsimony informative.
Species delimitation and diversity
The ABGD analysis of the 16S rRNA dataset retrieved 21 lineages when a barcode gap threshold of 1 4.9% was employed. The 21 lineages were designated to Prionospio sp. A to sp. P, Aurospio sp. Q to sp. T, as well as Aurospio cf. dibranchiata and Aurospio foodbancsia Mincks et al. 2009. The lineage Aurospio cf. dibranchiata is named according to corresponding GenBank records assigned in the analyses (Supplement 1), but with reservation, as no genetic data is available for the holotype or from the type locality. With higher barcode threshold values, a few lineages collapsed (5.1–5.2% = 20 lineages: lineages Prionospio sp. C and sp. D collapse; 5.4–8.5% = 19 lineages: lineages Prionospio sp. N and sp. O collapse). The GMYC analyses recovered also 21 lineages with slight differences to the results of the ABGD analysis. The lineages Prionospio sp. C and sp. D were grouped together into one lineage and lineage Prionospio sp. H was separated in two lineages.
Pairwise genetic distances (uncorrected p-distances) between the 21 lineages varied for 16S rRNA from 5.1–32.0% (based on the 21 lineages derived from 16S) (Table 2). The lowest pairwise distances were found between lineages Prionospio sp. C and sp. D (16S: 5.1–6.2%), as well as between lineages Prionospio sp. N and sp. O (16S: 5.2–5.9%; Table 2). All other inter-lineage distances exceeded 8.5% for 16S rRNA. The highest observed intra-lineage distances were found in lineage Prionospio sp. H with 4.8% for 16S rRNA (Table 2). Pairwise distances between the single lineages found for 18S varied from 0 to 1.9%, with highest distances between lineage Prionospio sp. E and sp. G and the highest intra-lineage pairwise distances were found for lineage Prionospio sp. E with 0.1% (Table 2).
The phylogenetic analyses of 16S recovered all 21 lineages as reciprocal monophyletic with full support each (Fig. 2; supplement 2). The two genera were not recovered as monophyletic as Aurospio foodbancsia, Aurospio sp. R and Aurospio sp. Q were nested among the Prionospio lineages (Fig. 2) and Aurospio sp. S, sp. T and Aurospio cf. dibranchiata formed a monophyletic clade, but also, this clade was nested within Prionospio lineages (Fig. 2). The approximately unbiased test rejected the ML topology, in which the genus Aurospio was constrained to be monophyletic, as being significantly worse than the unconstrained ML topology (p = 1.0 × 10−81).
Phylogenetic tree of Prionospio and Aurospio species obtained in the study based on mitochondrial 16S gene fragments. Individual specimens can be found in Supplement 2. Posterior probabilities shown next to the nodes (values below 0.8 are not shown). Bootstrap values are shown after slashes (values below 80 are not shown). Numbers of specimens are given in brackets. Individual occurrence in different oceans is color coded non proportional (see legend in the right upper corner)
In the haplotype networks of 16S rRNA (Fig. 3), a total of 128 haplotypes (h1-16S h128-16S) were found with a maximum of 28 haplotypes in one lineage (Prionospio sp. H, h31-16S h58-16S; Fig. 3). Notably, Prionospio H and Aurospio cf. dibranchiata had the highest intra-lineage 16S distances with up to 4.8%, and in both cases, these high distances were observed among individuals from Atlantic and Pacific oceans. However, other individuals from these regions either shared 16S rRNA haplotypes or featured haplotypes separated by only 1.3% (Fig. 3; Table 2).
The 18S network showed a total of 18 genotypes (h1-18S h18-18S; Fig. 4). The lineages Prionospio sp. D, sp. E, sp. G, sp. O and sp. P are well differentiated from each other, while other lineages shared genotypes and cluster together with no or one mutational step (Prionospio sp. L, sp. M, sp. N and Aurospio foodbancsia; Prionospio sp. H and sp. I; Prionospio sp. B and sp. C; Aurospio cf. dibranchiata and Aurospio sp. S; Fig. 4).
For several Prionospio and Aurospio lineages, pan-oceanic distributions with occurrences in the Atlantic as well as Pacific were recorded. Prionospio sp. A, sp. B, sp. E, sp. H, sp. L, and sp. N, as well as Aurospio sp. S and Aurospio cf. dibranchiata, were recorded from the VFZ in the Atlantic and the CCZ in the Pacific whereas Aurospio cf. dibranchiata and Prionospio sp. E occurred furthermore in the Southern Ocean (Fig. 2; Supplement 2). However, not only lineages, but also five 16S rRNA haplotypes were shared between specimens from the Pacific and the Atlantic (Prionospio sp. B, h9-16S; Prionospio sp. H, h35-16S; Prionspio sp. L, h62-16S; Aurospio sp. S, h94-16S; Aurospio cf. dibranchiata, h111-16S; Fig. 3). The majority of the pan-oceanic distributed species show one haplotype per species shared between the Atlantic and the Pacific and a separation between both oceans is observable, though often by only few mutational steps. In Prionospio sp. A and sp. N specimens of each ocean do not share haplotypes and Atlantic and Pacific haplotypes are separated by three and six mutations, respectively (Fig. 3). Also for Prionospio sp. E, the haplotypes are not shared between oceans, but there is also no clear separation between Pacific and Atlantic as the haplotypes of the Atlantic intermingle with haplotypes from the Pacific (and to a lesser degree with those from Antarctica) (Fig. 3). The single specimen from the Pacific assigned to Prionospio sp. L shares a haplotype with specimens from the Atlantic (Fig. 3; Supplement 1; h62-16S: 1 Pacific specimen and 6 Atlantic specimens). Prionospio sp. B show a pan-oceanic shared haplotype (Fig. 3; Supplement 1; h9-16S: three Atlantic specimens and nine Pacific specimens) and no obvious separation of the oceans. Population genetics of Prionospio sp. B showed significant difference between the population from the eVFZ and the population from the Pacific, but no significant differences between the specimens from the VTF and the Pacific were found (Table 4). Also, Prionospio sp. H and Aurospio cf. dibranchiata each have one haplotype distributed across the Atlantic and the Pacific (Fig. 3; Supplement 1; h35-16S: one Pacific and one Atlantic specimen; h118-16S: one Pacific and 23 Atlantic specimens), and a separation of both oceans can be seen in the networks with a minimum of four mutations in Prionospio sp. H and two mutations in Aurospio cf. dibranchiata between the other haplotypes. Population differentiation was significant between the Atlantic and Pacific specimens for Prionospio sp. H (Table 4). For Aurospio cf. dibranchiata, significant differences were also found within the Atlantic population (eVFZ and VTF; Table 4). Tajima’s D and Fu’s Fs reveal significant negative values for Prionospio sp. H (Table 4). Aurospio cf. dibranchiata showed also negative significant values, but only for the Atlantic population (Table 3: Fu’s Fs). Aurospio sp. S showed one shared haplotype (Fig. 3; Supplement 1; h94-16S: one Pacific and four Atlantic specimens), but with no clear separation of both oceans, as most other haplotypes are separated by only one mutational step from the shared haplotype. Nevertheless, population differentiation was significant between the Atlantic and Pacific specimens for lineage Aurospio sp. S (Table 4).
Thirteen of the identified lineages were restricted to one of the sampled oceans: Prionospio sp. D, sp. G, sp. I, sp. M, sp. O and sp. P to the Atlantic; Prionospio sp. C, sp. F, sp. K and Aurospio sp. T to the Pacific; Aurospio foodbancsia and Aurospio sp. R to the Southern Ocean; and Aurospio sp. Q to the Arabian Sea (Fig. 2). Moreover, some of these lineages were distributed over larger regional scales within these oceans and sea. For instance, Prionospio sp. G had one haplotype recorded from the eVFZ and the wVFZ (Fig. 3: h29-16S) and Prionospio sp. L even shared a haplotype between the eVFZ and the PRT (Fig. 3: h61-16S).
Discussion
In the present study based on the mitochondrial marker 16S rRNA gene, 21 lineages were identified consistently and well supported for Prionospio and Aurospio from the deep Atlantic and Pacific. But limits of mitochondrial markers for species delimitation are well known, as mitochondrial DNA poses potential risks like the retention of ancestral polymorphism, male-biased gene flow, hybrid introgression, and paralogy (Galtier et al. 2009; Moritz and Cicero 2004). Species delimitation with GMYC is largely corresponding to the lineages identified with ABGD, except the collapse of lineages Prionospio sp. C and sp. D to one lineage and the separation of lineage Prionospio sp. H into two lineages. However, taking into account the results for the nuclear 18S gene, the lineages Prionospio sp. D, sp. E, sp. G, sp. O and sp. P can be easily differentiated. Other lineages share 18S genotypes (Prionospio sp. B and sp. C; Prionospio sp. H and sp. I; Prionospio sp. L, sp. M, sp. N and Aurospio foodbancsia; Aurospio cf. dibranchiata and Aurospio sp. S), but lacking differentiation in 18S analyses is not surprising, as the gene is known for the lower discriminating power at species level for metazoans (Halanych and Janosik 2006; Hillis and Dixon 1991). The lack of differentiation for some lineages in 18S is probably not the result of the lack of reproductive isolation among these species, but rather caused by a combination of the lower substitution rate in this marker and incomplete lineage sorting (Funk and Omland 2003). All lineages, which share an identical 18S genotype, had uncorrected pairwise distances exceeding 8.4% in 16S rRNA (lineages Prionospio sp. B and sp. C: 8.4–10.3%; lineages Prionospio sp. H and sp. I: 11.6–16.6%; lineages Prionospio sp. L, sp. M, sp. N and Aurospio foodbancsia: 12.8–22.9%; lineages Aurospio cf. dibranchiata and sp. S: 13.2–16.9%) and the intraspecific uncorrected distances were always lower within these lineages (16S: 0–5.8%). The levels of interspecific differentiation in 16S rRNA are comparable to those observed among other polychaete species, which usually exceeded 5% for the mitochondrial marker 16S (Álvarez-Campos et al. 2017; Brasier et al. 2016; Meißner et al. 2016; Wiklund et al. 2009). We imply thresholds about 5–6% for 16S rRNA to distinguish between Prionospio and Aurospio species. In a recent study, Guggolz et al. (2019) found barcoding gaps of ~ 2–2.8% in 16S rRNA between Laonice species, another spionid genus, from the tropical Atlantic. The fact that we found higher intraspecific distances, in particular within Prionospio sp. H and Aurospio cf. dibranchiata, might be explained by the larger geographic sampling in our study. It appears likely that the overall intraspecific diversity would be higher if additional regions and populations were studied. Based on the higher pairwise distances for 16S rRNA between lineages C and D (5.1–6.2%) compared to the distances within lineage Prionospio sp. H (4.8%), we follow the species delimitation of ABGD instead of GMYC. Based on the available data, we postulate that each one of the 21 lineages, identified with ABGD, represents a separate species within the investigated dataset with reservations for lineages Prionospio sp. C, sp. D and sp. H. One has to keep in mind that a higher number of species might be present within the dataset (e.g., Prionospio sp. E and sp. A might be further split) using a less conservative threshold for delimitating species, but this additional splitting and the differences found between ABGD and GMYC analyses would not change the main finding, concerning the dispersal potential of Prionospio and Aurospio (see below).
Unfortunately, our previous morphology-based species identification of the Prionospio and Aurospio specimens from the tropical Atlantic (Guggolz et al. 2018) showed high inconsistencies with the results presented here regarding the number of species. The assignment to genera showed consistency with the molecular results, but the number of species had been underestimated with morphological identification (Guggolz et al. 2018: eight Prionospio species, one Aurospio species). As all of the studied specimens were damaged and important species discriminating characters were often missing, morphological identification was limited, which could explain this inconsistency. The problem of morphological identification is known for deep-sea polychaetes, due to their soft bodies resulting in easy fragmentation and the way of sampling infauna from deep-sea sediments, where extreme care must be taken regarding sieving techniques to preserve morphology (Bogantes et al. 2018; Glover et al. 2016; Guggolz et al. 2019). Furthermore, Prionospio and Aurospio are classified within the Prionospio complex, in which the generic characters are limited and still under debate (Paterson et al. 2016; Sigvaldadottir 1998; Wilson 1990; Yokoyama 2007). Especially, the taxonomic boundaries of Prionospio and Aurospio are problematic, as the main distinguishing feature is the beginning and shape of the branchiae, which has been suggested to be insufficient to discriminate the genera (Paterson et al. 2016; Sigvaldadottir 1998; Wilson 1990). The 16S rRNA phylogeny of the present work highlights this difficulty as monophyly for both genera, Prionospio and Aurospio, was not supported. Of the 21 species identified within the study, Aurospio foodbancsia, Aurospio, sp. Q and sp. R were studied solely on the basis of sequence data obtained from GenBank. For Aurospio sp. Q and sp. R, only single sequences each were available from GenBank without further taxonomic information and these could have been misidentified, a well-known problem (Collins and Cruickshank 2012; Kvist et al. 2010). In contrast, the available sequences for Aurospio foodbancsia are based on the original description of the type specimen (Mincks et al. 2009). However, even if a misidentification of Aurospio sp. R and sp. Q was assumed, the relationship of Aurospio foodbancsia does not support monophyly of Aurospio. Aurospio cf. dibranchiata, A. sp. S and sp. T cluster within Prionospio in the 16S rRNA analyses and shared a 18S genotype with Prionospio species also the monophyly of Prionospio is not supported. Although we could not find support for the monophyly of the two genera, conclusions about the paraphyly of both genera should be treated with caution, as the available 16S rRNA and 18S data might not sufficiently reflect relationships. To resolve these problems, there is a need for broad taxon sampling and a taxonomic revision including morphological and genetic data including more genes.
Irrespective of the taxonomic status of Prionospio and Aurospio, the dataset presented here provides profound insights into the diversity of these genera in the deep sea. In the samples for the tropical Atlantic studied herein, spionids were a dominant faunal component with more than 73% of all sampled polychaetes (Guggolz et al. 2018). The potential 19 deep-sea species (excluding the two GenBank entries Aurospio sp. Q and R) make up around 17% of the worldwide described Prionospio and Aurospio species (around 108 species listed in WoRMS http://www.marinespecies.org/, Read and Fauchald 2018), keeping in mind that we cannot say for sure whether they represent species new to science or already described species. Our findings support previous reports that both genera are abundant in the deep sea, and presumably the number of species is still underestimated (Paterson et al. 2016).
One of the main aims of this study was to assess the distribution and dispersal potential of the studied species. Some species were found to be restricted to one of the investigated oceans (Prionospio sp. C, D, F, G, I, K, L, M, O, P and Aurospio sp. T; Figs. 2 and 3), but there are also several species found to have a pan-oceanic distribution (Prionospio sp. A, B, E, H, N and Aurospio cf. dibranchiata, sp. S; Figs. 2 and 3). Most of these species found in the Atlantic, as well as in the Pacific, show comparable patterns regarding the haplotype networks and thus of the distribution of their intraspecific genetic diversity. Identical haplotypes are shared between populations inhabiting both oceans (i.e., h9-16S, h35-16S, h95-16S, h119-16S; Fig. 3), and other haplotypes differ only on one or a few mutations in 16S rRNA between oceans. There are three possible scenarios explaining this remarkable finding: (1) shared haplotypes were already present in the ancestral population > 3 million years ago (O’Dea et al. 2016) before Atlantic and Pacific populations were separated by the Isthmus of Panama (vicariance), (2) historic gene flow between Atlantic and Pacific populations, and (3) recent and/or continuous gene flow between Atlantic and Pacific populations. In scenario (1), the observed distribution of haplotypes would be due to variance rather than dispersal and gene flow. Identical haplotypes would be ancestral and retained unchanged in both populations over 3 million years. This would imply very low substitution and speciation rates for these species as we have no reason to believe that the Atlantic and Pacific populations in question represent different species. In turn, this would imply very old ages for the studied genera and all other studied species pairs, as these species were separated by 5.1–32.0% uncorrected p-distance in 16S rRNA, which would require hundreds of millions of years to accumulate at this rate. We deem this rather unlikely and therefore favor scenarios (2) and (3).
Scenarios (2) and (3) both assume gene flow between Atlantic and Pacific populations of several species much later than the closure of the Isthmus of Panama. With the available data, it is not possible to assess the timing and frequency of gene flow events within each species. However, dispersal and gene flow rates are apparently too low to lead to population admixture between oceans. In several of the studied species, dispersal might have occurred only once, resulting in the colonization of the respectively other ocean, though repeated and on-going dispersal and gene flow is possible as well.
It is widely accepted that one of the main driving factors for such wide dispersal capacities of marine invertebrates are long-lived planktonic larval stages (Eckman 1996; Rex et al. 2005; Scheltema 1972; Schüller and Ebbe 2007; Yearsley and Sigwart 2011). Even if the specific type of larvae of the herein studied species is unknown, other species of Prionospio and Aurospio are reported to develop via planktonic larvae like other species of these genera (Blake et al. 2018; Mincks et al. 2009; Young 2003). The planktonic larval duration and dispersal distances are potentially higher in the deep sea, as the cold temperature and consequently reduced metabolic rates are identified as one of the main driving factors for extended larval stages (McClain and Hardy 2010; O’Connor et al. 2007). Considering the enormous distances between the CCZ and the VFZ, it seems highly unlikely that larvae drift all the way between the populations. It rather appears plausible that the distribution over such large geographical distances is linked to ocean currents, connecting different suitable habitat patches in a stepwise fashion (McClain and Hardy 2010; Rex and Etter 2010; Young et al. 2008). The direction of dispersal of Aurospio and Prionospio larvae remains unknown, as well as whether the species originated from the Pacific or the Atlantic. Probably, additional populations occur in the un-sampled regions between our study sites, which might have acted as stepping stones. Alternatively, some of the studied species might have a continuous distribution linking the Atlantic and Pacific populations. Further comparison to populations from other localities would help to clarify these issues. The potential to find conspecific specimens in other deep-sea areas and another indication for the enormous distribution potential of some spionids is supported by specimens from the Crozet Islands (Antarctica) that were assigned to Prionospio sp. E. The Southern Ocean could also connect the Atlantic and the Pacific most likely with the eastwards flowing deep-water currents (Rahmstorf 2002; Stow et al. 2002).
A pan-oceanic distribution found for Aurospio and Prionospio species as evidenced from molecular data is so far unique for abyssal annelids, even if wide distribution ranges of polychaete species are not unusual (Böggemann 2016; Eilertsen et al. 2018; Georgieva et al. 2015; Guggolz et al. 2019; Meißner et al. 2016; Schüller and Hutchings 2012). However, Hutchings and Kupriyanova (2018) are particularly highlighting that reported “cosmopolitan” species should be treated with caution. For some species, wide distribution ranges have been based on misidentification or cryptic species and subsequently rejected using molecular marker (Álvarez-Campos et al. 2017; Nygren 2014; Nygren et al. 2018; Sun et al. 2016). The assumption that planktonic larvae in the deep sea are staying longer in the water column and transported via currents successfully over long distances raises the question of what the potential dispersal barrier restricting the distribution are. Life-history traits like larval behavior, larval mortality, and physiological tolerances for vertical movement and settlement in different habitats are probably important (Glover et al. 2001; McClain and Hardy 2010; Virgilio et al. 2009). It has been suggested that connectivity in the deep sea has often been associated with a common bathymetry rather than spatial vicinity (Glover et al. 2001). Thus, topographic barriers, like ridges, canyons, and rises, are supposed to influence distribution patterns (Eckman 1996; Guggolz et al. 2018; McClain and Hardy 2010; Won et al. 2003). Such a potential barrier is the MAR, dividing the Atlantic in eastern and western basins (Riehl et al. 2018). Some of the herein studied species (Prionospio sp. G, sp. L, sp. M and Aurospio cf. dibranchiata) occurring in the tropical Atlantic were found to be distributed across the MAR with wide distribution ranges of up to > 4000 km (Figs. 1 and 3) with no or only little genetic differentiation between populations. Several haplotypes were found to be identical (h29-16S, h59-16S, h61-16S, h69-16S, h119-16S, h121-16S; Fig. 3), indicating gene flow across the MAR. Even for species restricted to one side of the MAR (eVFZ: Prionospio sp. H, sp. P and Aurospio sp. S) or at least to the VTF (Prionspio sp. B, sp. E, sp. N, sp. O), shared haplotypes were found across hundreds of kilometers (47-16S, h9-16S, h22-16S, h87-16S; Figs. 1 and 3). These results are strongly refuting a barrier effect of the MAR for these Prionospio and Aurospio species. The dispersal potential across topographic barriers like the MAR and over large geographic distances was also reported recently for species of the spionid genus Laonice (Guggolz et al. 2019). The importance of dispersal via larvae is emphasized by distribution patterns found for brooding taxa like isopods, which exhibited very limited or no gene flow across the MAR (Bober et al. 2018; Brix et al. 2018).
In summary, the results of this study are expanding our knowledge of the diversity and distribution patterns of Aurospio and Prionospio in the deep sea. We could identify 21 lineages with some assigned to already described species (Aurospio foodbancsia, Aurospio cf. dibranchiata) and others remaining unknown at present. These other molecularly delineated potential species cannot be differentiated from described species, due to lack of sufficient morphological characters as most individuals were damaged and incomplete and because genetic data for most of the described species is lacking. The data do not support the monophyly for the two genera, which is highlighting the importance of molecular analyses additionally to morphological examinations to revise the Prionospio complex. A remarkable degree of pan-oceanic distribution in some of the species was shown, especially in the tropical Atlantic. The pan-oceanic distribution is indicating the potential of widespread distribution for Prionospio and Aurospio species, even over topographic barriers and should be kept in mind for further studies on distribution patterns in deep-sea polychaetes.
Data availability
The datasets generated during and/or analyzed during the current study are available in the GenBank repository (see corresponding GenBank numbers in Supplementary file 2).
References
Álvarez-Campos, P., Giribet, G., & Riesgo, A. (2017). The Syllis gracilis species complex: a molecular approach to a difficult taxonomic problem (Annelida, Syllidae). Molecular Phylogenetics and Evolution, 109, 138–150. https://doi.org/10.1016/j.ympev.2016.12.036.
Bandelt, H. J., Forster, P., & Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16(1), 37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036.
Bickford, D., Lohman, D. J., Sodhi, N. S., Ng, P. K. L., Meier, R., Winker, K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends in Ecology & Evolution, 22(3), 148–155. https://doi.org/10.1016/j.tree.2006.11.004.
Blake, J. A., & Kudenov, J. D. (1978). The Spionidae (Polychaeta) from southeastern Australia and adjacent areas with a revision of the genera. Memoirs of the National Museum of Victoria, 39, 171–280.
Blake, J. A., Maciolek, N. J., & Meißner, K. (2018). Handbook of Zoology: Spionidae Grube, 1850. In A Natural History of the Phyla of the Animal Kingdom, Annelida: Polychaeta (p. 109 pp). Berlin, Boston.
Bober, S., Brix, S., Riehl, T., Schwentner, M., & Brandt, A. (2018). Does the mid-Atlantic ridge affect the distribution of abyssal benthic crustaceans across the Atlantic Ocean? Deep Sea Research Part II: Topical Studies in Oceanography, 148, 91–104. https://doi.org/10.1016/j.dsr2.2018.02.007.
Bogantes, V. E., Halanych, K. M., & Meißner, K. (2018). Diversity and phylogenetic relationships of North Atlantic Laonice Malmgren, 1867 (Spionidae, Annelida) including the description of a novel species. Marine Biodiversity, 48(2), 737–749. https://doi.org/10.1007/s12526-018-0859-8.
Böggemann, M. (2016). Glyceriformia (Annelida) of the abyssal SW Atlantic and additional material from the SE Atlantic. Marine Biodiversity, 46(1), 227–241. https://doi.org/10.1007/s12526-015-0354-4.
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.-H., Xie, D., Suchard, M. A., Rambaut, A., & Drummond, A. J. (2014). BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 10(4), e1003537. https://doi.org/10.1371/journal.pcbi.1003537.
Brasier, M. J., Wiklund, H., Neal, L., Jeffreys, R., Linse, K., Ruhl, H., & Glover, A. G. (2016). DNA barcoding uncovers cryptic diversity in 50% of deep-sea Antarctic polychaetes. Royal Society Open Science, 3(11), 160432. https://doi.org/10.1098/rsos.160432.
Brix, S., Bober, S., Tschesche, C., Kihara, T.-C., Driskell, A., & Jennings, R. M. (2018). Molecular species delimitation and its implications for species descriptions using desmosomatid and nannoniscid isopods from the VEMA fracture zone as example taxa. Deep Sea Research Part II: Topical Studies in Oceanography, 148, 180–207. https://doi.org/10.1016/j.dsr2.2018.02.004.
Caullery, M. (1914). Sur les polychètes du genre Prionospio Malmgren. Bulletin de le Soci zoologique de France, 39, 355–361.
Cohen B.L., Gawthrop A., Cavalier-Smith T. (1998). Molecular phylogeny of brachiopods and phoronids based on nuclear-encoded small subunit ribosomal RNA gene sequences. Philosophical Transactions of the Royal Society B Biological Sciences, 353, 2039–2061.
Collins, R. A., & Cruickshank, R. H. (2012). The seven deadly sins of DNA barcoding. Molecular Ecology Resources, n/a-n/a. https://doi.org/10.1111/1755-0998.12046.
Dahlgren, T. G., Wiklund, H., Rabone, M., Amon, D. J., Ikebe, C., Watling, L., et al. (2016). Abyssal fauna of the UK-1 polymetallic nodule exploration area, Clarion-Clipperton Zone, central Pacific Ocean: Cnidaria. Biodiversity Data Journal, 4. https://doi.org/10.3897/BDJ.4.e9277.
Devey, C. W. (2015). RV SONNE Fahrtbericht / Cruise Report SO237 Vema-TRANSIT : bathymetry of the Vema-Fracture-Zone and Puerto Rico TRench and Abyssal AtlaNtic BiodiverSITy Study, Las Palmas (Spain) - Santo Domingo (Dom. Rep.) 14.12.14–26.01.15. doi:Devey, Colin W., ed. and Shipboard scientific party (2015) RV SONNE Fahrtbericht / Cruise Report SO237 Vema-TRANSIT : bathymetry of the Vema-Fracture-Zone and Puerto Rico TRench and Abyssal AtlaNtic BiodiverSITy Study, Las Palmas (Spain) - Santo Domingo (Dom. Rep.) 14.12.14–26.01.15. Open Access. GEOMAR Report, N. Ser. 023. GEOMAR Helmholtz-Zentrum für Ozeanforschung Kiel, Kiel, Germany, 130 pp.
Dzikowski, R., Levy, M.G., Poore, M.F., Flowers, J.R., Paperna, I. (2004). Use of rDNA polymorphism for identification of Heterophyidae infecting freshwater fishes. Diseases of Aquatic Organisms 59(1), 35–41.
Eckman, J. E. (1996). Closing the larval loop: linking larval ecology to the population dynamics of marine benthic invertebrates. Journal of Experimental Marine Biology and Ecology, 200(1–2), 207–237. https://doi.org/10.1016/S0022-0981(96)02644-5.
Eilertsen, M. H., Georgieva, M. N., Kongsrud, J. A., Linse, K., Wiklund, H., Glover, A. G., & Rapp, H. T. (2018). Genetic connectivity from the Arctic to the Antarctic: Sclerolinum contortum and Nicomache lokii (Annelida) are both widespread in reducing environments. Scientific Reports, 8(1). https://doi.org/10.1038/s41598-018-23076-0.
Etter, R. J., Boyle, E. E., Glazier, A., Jennings, R. M., Dutra, E., & Chase, M. R. (2011). Phylogeography of a pan-Atlantic abyssal protobranch bivalve: implications for evolution in the Deep Atlantic. Molecular Ecology, 20(4), 829–843. https://doi.org/10.1111/j.1365-294X.2010.04978.x.
Excoffier, L., & Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10(3), 564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x.
Foster, N. M. (1971). Spionidae (Polychaeta) of the Gulf of Mexico and the Caribbean Sea. The Hague.
Funk, D. J., & Omland, K. E. (2003). Species-lefvel paraphyly and polyphyly: drequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics, 34(1), 397–423. https://doi.org/10.1146/annurev.ecolsys.34.011802.132421.
Galtier, N., Nabholz, B., GléMin, S., & Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology, 18(22), 4541–4550. https://doi.org/10.1111/j.1365-294X.2009.04380.x.
Georgieva, M. N., Wiklund, H., Bell, J. B., Eilertsen, M. H., Mills, R. A., Little, C. T. S., & Glover, A. G. (2015). A chemosynthetic weed: the tubeworm Sclerolinum contortum is a bipolar, cosmopolitan species. BMC Evolutionary Biology, 15(1), 1–17. https://doi.org/10.1186/s12862-015-0559-y.
Glover, A. G., & Fauchald, K. (2018). World Polychaeta database. Prionospio Malmgren, 1867. Accessed through: Glover, A.G., Higgs, N., Horton, T. World Register of Deep-Sea species.
Glover, A., Paterson, G., Bett, B., Gage, J., Sibuet, M., Sheader, M., & Hawkins, L. (2001). Patterns in polychaete abundance and diversity from the Madeira Abyssal Plain, northeast Atlantic. Deep Sea Research Part I: Oceanographic Research Papers, 48(1), 217–236. https://doi.org/10.1016/S0967-0637(00)00053-4.
Glover, A., Dahlgren, T. G., Wiklund, H., Mohrbeck, I., & Smith, C. (2016). An end-to-end DNA taxonomy methodology for benthic biodiversity survey in the Clarion-Clipperton Zone, Central Pacific abyss. Journal of Marine Science and Engineering, 4(1), 2. https://doi.org/10.3390/jmse4010002.
Guggolz, T., Lins, L., Meißner, K., & Brandt, A. (2018). Biodiversity and distribution of polynoid and spionid polychaetes (Annelida) in the Vema Fracture Zone, tropical North Atlantic. Deep Sea Research Part II: Topical Studies in Oceanography, 148, 54–63. https://doi.org/10.1016/j.dsr2.2017.07.013.
Guggolz, T., Meißner, K., Schwentner, M., & Brandt, A. (2019). Diversity and distribution of Laonice species (Annelida: Spionidae) in the tropical North Atlantic and Puerto Rico Trench. Scientific Reports, 9(1), 9260. https://doi.org/10.1038/s41598-019-45807-7.
Halanych, K. M., & Janosik, A. M. (2006). A review of molecular markers used for Annelid phylogenetics. Integrative and Comparative Biology, 46(4), 533–543. https://doi.org/10.1093/icb/icj052.
Hillis, D. M., & Dixon, M. T. (1991). Ribosomal DNA: molecular evolution and phylogenetic inference. The Quarterly Review of Biology, 66(4), 411–453. https://doi.org/10.1086/417338.
Hutchings, P., & Kupriyanova, E. (2018). Cosmopolitan polychaetes – fact or fiction? Personal and historical perspectives. Invertebrate Systematics, 32(1), 1. https://doi.org/10.1071/IS17035.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. https://doi.org/10.1093/molbev/mst010.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P., & Drummond, A. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. https://doi.org/10.1093/bioinformatics/bts199.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. https://doi.org/10.1093/molbev/msw054.
Kvist, S., Oceguera-Figueroa, A., Siddall, M. E., & Erséus, C. (2010). Barcoding, types and the Hirudo files: using information content to critically evaluate the identity of DNA barcodes. Mitochondrial DNA, 21(6), 198–205. https://doi.org/10.3109/19401736.2010.529905.
Linse, K., & Schwabe, E. (2018). Diversity of macrofaunal Mollusca of the abyssal Vema Fracture Zone and hadal Puerto Rico Trench, Tropical North Atlantic. Deep Sea Research Part II: Topical Studies in Oceanography, 148, 45–53. https://doi.org/10.1016/j.dsr2.2017.02.001.
Lodge, M., Johnson, D., Le Gurun, G., Wengler, M., Weaver, P., & Gunn, V. (2014). Seabed mining: international seabed authority environmental management plan for the Clarion–Clipperton Zone. A partnership approach. Marine Policy, 49, 66–72. https://doi.org/10.1016/j.marpol.2014.04.006.
Maciolek, N. J. (1981). A new genus and species of Spionidae (Annelida:Polychaeta) from the North and South Atlantic. Proceedings of the Biological Society of Washington, 94(1), 12.
Maciolek, N. J. (1981a). Spionidae (Annelida: Polychaeta) from the Galapagos Rift geothermal vents. Proceedings of the Biological Society of Washington, 94(3), 826–837.
Maciolek, N. J. (1985). A revision of the genus Prionospio Malmgren, with special emphasis on species from the Atlantic Ocean, and new records of species belonging to the genera ApoPrionospio Foster and Paraprionospio Caullery (Polychaeta, Annelida, Spionidae). Zoological Journal of the Linnean Society, 84(4), 325–383. https://doi.org/10.1111/j.1096-3642.1985.tb01804.x.
Malmgren, A. J. (1867). Annulata polychaeta: Spetsbergiæ, Grœnlandiæ, Islandiæ et Scandinaviæ. Ex Officina Frenckelliana: Hactenus cognita.
Martínez Arbizu, P., & Haeckel, M. (2015). RV SONNE Fahrtbericht / Cruise Report SO239: EcoResponse Assessing the ecology, connectivity and resilience of polymetallic nodule field systems, Balboa (Panama) – Manzanillo (Mexico,) 11.03.-30.04.2015. GEOMAR Helmholtz-Zentrum für Ozeanforschung Kiel.
McClain, C. R., & Hardy, S. M. (2010). The dynamics of biogeographic ranges in the deep sea. Proceedings of the Royal Society B: Biological Sciences, 277(1700), 3533–3546. https://doi.org/10.1098/rspb.2010.1057.
McClain, C. R., Rex, M. A., & Etter, R. J. (2009). Deep-sea macroecology. In Marine Macroecology (pp. 65–100). Chicago: University of Chicago Press.
Medlin L., Elwood H.J., Stickel S., Sogin M.L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene, 71, 491–499.
Meißner, K., & Blank, M. (2009). Spiophanes norrisi sp. nov. (Polychaeta: Spionidae) a new species from the NE Pacific coast, separated from the Spiophanes bombyx complex based on both morphological and genetic studies. Zootaxa, 2278(1), 1–25.
Meißner, K., Bick, A., Guggolz, T., & Götting, M. (2014). Spionidae (Polychaeta: Canalipalpata: Spionida) from seamounts in the NE Atlantic. Zootaxa, 3786, 201–245. https://doi.org/10.11646/zootaxa.3786.3.1.
Meißner, K., Bick, A., & Götting, M. (2016). Arctic Pholoe (Polychaeta: Pholoidae): when integrative taxonomy helps to sort out barcodes. Zoological Journal of the Linnean Society. https://doi.org/10.1111/zoj.12468.
Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees. In 2010 Gateway Computing Environments Workshop (GCE) (pp. 1–8). Presented at the 2010 Gateway Computing Environments Workshop (GCE). doi:https://doi.org/10.1109/GCE.2010.5676129.
Mincks, S. L., Dyal, P. L., Paterson, G. L. J., Smith, C. R., & Glover, A. G. (2009). A new species of Aurospio (Polychaeta, Spionidae) from the Antarctic shelf, with analysis of its ecology, reproductive biology and evolutionary history. Marine Ecology, 30(2), 181–197. https://doi.org/10.1111/j.1439-0485.2008.00265.x.
Moritz, C., & Cicero, C. (2004). DNA barcoding: promise and pitfalls. PLoS Biology, 2(10), e354. https://doi.org/10.1371/journal.pbio.0020354.
Nygren, A. (2014). Cryptic polychaete diversity: a review. Zoologica Scripta, 43(2), 172–183. https://doi.org/10.1111/zsc.12044.
Nygren A., Sundberg P. (2003) Phylogeny and evolution of reproductive modes in Autolytinae (Syllidae, Annelida). Molecular Phylogenetics and Evolution, 29, 235–249.
Nygren, A., Parapar, J., Pons, J., Meißner, K., Bakken, T., Kongsrud, J. A., Oug, E., Gaeva, D., Sikorski, A., Johansen, R. A., Hutchings, P. A., Lavesque, N., & Capa, M. (2018). A mega-cryptic species complex hidden among one of the most common annelids in the North East Atlantic. PLoS One, 13(6), e0198356. https://doi.org/10.1371/journal.pone.0198356.
O’Connor, M. I., Bruno, J. F., Gaines, S. D., Halpern, B. S., Lester, S. E., Kinlan, B. P., & Weiss, J. M. (2007). Temperature control of larval dispersal and the implications for marine ecology, evolution, and conservation. Proceedings of the National Academy of Sciences, 104(4), 1266–1271. https://doi.org/10.1073/pnas.0603422104.
O’Dea, A., Lessios, H. A., Coates, A. G., Eytan, R. I., Restrepo-Moreno, S. A., Cione, A. L., et al. (2016). Formation of the isthmus of Panama. Science Advances, 2(8), e1600883. https://doi.org/10.1126/sciadv.1600883.
Palumbi, S. R. et al. (1991). The Simple Fool's Guide to PCR, Version 2.0. Privately published, Univ. Hawaii.
Palumbi, S.R. (1996). Nucleic acids II: the polymerase chain reaction. In: Molecular Systematics (pp. 205–247). Massachusetts: Sinauer &Associates Inc., Sunderland.
Paterson, G. L. J., Neal, L., Altamira, I., Soto, E. H., Smith, C. R., Menot, L., et al. (2016). New Prionospio and Aurospio species from the deep sea (Annelida: Polychaeta). Zootaxa, 4092(1), 1. https://doi.org/10.11646/zootaxa.4092.1.1.
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., Kamoun, S., Sumlin, W. D., & Vogler, A. P. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology, 55(4), 595–609. https://doi.org/10.1080/10635150600852011.
Priede, I. G., Bergstad, O. A., Miller, P. I., Vecchione, M., Gebruk, A., Falkenhaug, T., Billett, D. S., Craig, J., Dale, A. C., Shields, M. A., Tilstone, G. H., Sutton, T. T., Gooday, A. J., Inall, M. E., Jones, D. O., Martinez-Vicente, V., Menezes, G. M., Niedzielski, T., Sigurðsson, Þ., Rothe, N., Rogacheva, A., Alt, C. H., Brand, T., Abell, R., Brierley, A. S., Cousins, N. J., Crockard, D., Hoelzel, A. R., Høines, Å., Letessier, T. B., Read, J. F., Shimmield, T., Cox, M. J., Galbraith, J. K., Gordon, J. D., Horton, T., Neat, F., & Lorance, P. (2013). Does presence of a mid-ocean ridge enhance biomass and biodiversity? PLoS One, 8(5), e61550. https://doi.org/10.1371/journal.pone.0061550.
Puillandre, N., Lambert, A., Brouillet, S., & Achaz, G. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation: ABGD, Automatic Barcode Gap Discovery. Molecular Ecology, 21(8), 1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x.
R Core Team. (2018). R: a language and environment for statistical computing. Vienna: R foundation for statistical computing.
Rahmstorf, S. (2002). Ocean circulation and climate during the past 120,000 years. Nature, 419(6903), 207–214. https://doi.org/10.1038/nature01090.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., & Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67(5), 901–904. https://doi.org/10.1093/sysbio/syy032.
Read, G., Fauchald, K. (Ed.) (2018). World Polychaeta database. Prionospio Malmgren, 1867. Aurospio Maciolek, 1981. Accessed through: World Register of Marine Species at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=129620. Accessed 11 February 2020.
Rex, M. A., & Etter, R. J. (2010). Deep-seabBiodiversity: pattern and scale. Harvard University Press.
Rex, M. A., McClain, C. R., Johnson, N. A., Etter, R. J., Allen, J. A., Bouchet, P., & Warén, A. (2005). A source-sink hypothesis for abyssal biodiversity. The American Naturalist, 165(2), 163–178. https://doi.org/10.1086/427226.
Riehl, T., Kaiser, S., & Brandt, A. (2018). Vema-TRANSIT–an interdisciplinary study on the bathymetry of the Vema-Fracture Zone and Puerto Rico Trench as well as abyssal Atlantic biodiversity. Deep Sea Research Part II: Topical Studies in Oceanography, 148, 1–6. https://doi.org/10.1016/j.dsr2.2018.01.007.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12), 1572–1574. https://doi.org/10.1093/bioinformatics/btg180.
Scheltema, R. S. (1972). Reproduction and dispersal of bottom dwelling deep-sea invertebrates: a speculative summary. In R. W. Brauer (Ed.), Barobiology and the experimental biology of the Deep Sea (pp. 58–66). Chapel Hill: University of North Carolina.
Schüller, M., & Ebbe, B. (2007). Global distributional patterns of selected deep-sea Polychaeta (Annelida) from the Southern Ocean. Deep Sea Research Part II: Topical Studies in Oceanography, 54(16–17), 1737–1751. https://doi.org/10.1016/j.dsr2.2007.07.005.
Schüller, M., & Hutchings, P. A. (2012). New species of Terebellides (Polychaeta: Trichobranchidae) from the deep Southern Ocean, with a key to all described species. Zootaxa, 3619(1), 1–31. https://doi.org/10.11646/zootaxa.3619.1.1.
Shimodaira, H. (2002). An approximately unbiased test of phylogenetic tree selection. Systematic Biology, 51, 492–508.
Shimodaira, H., & Hasegawa, M. (2001). CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics, 17, 1246–1247.
Sigvaldadottir, E. (1998). Cladistic analysis and classification of Prionospio and related general (Polychaeta, Spionidae). Zoologica Scripta, 27(3), 175–187. https://doi.org/10.1111/j.1463-6409.1998.tb00435.x.
Sigvaldadóttir, E., & Mackie, A. S. Y. (1993). Prionospio steenstrupi, P. fallax and P. dubia (Polychaeta, Spionidae): re-evaluation of identity and status. Sarsia, 78(3–4), 203–219. https://doi.org/10.1080/00364827.1993.10413535.
Sjölin, E., Erséus, C., & Källersjö, M. (2005). Phylogeny of Tubificidae (Annelida, Clitellata) based on mitochondrial and nuclear sequence data. Molecular Phylogenetics and Evolution, 35(2), 431–441.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. https://doi.org/10.1093/bioinformatics/btu033.
Stow, D. A. V., Faugères, J.-C., Howe, J. A., Pudsey, C. J., & Viana, A. R. (2002). Bottom currents, contourites and deep-sea sediment drifts: current state-of-the-art. Geological Society, London, Memoirs, 22(1), 7–20. https://doi.org/10.1144/GSL.MEM.2002.022.01.02.
Sun, Y., Wong, E., Tovar-Hernández, M. A., Williamson, J. E., & Kupriyanova, E. K. (2016). Is Hydroides brachyacantha (Serpulidae : Annelida) a widespread species? Invertebrate Systematics, 30(1), 41. https://doi.org/10.1071/IS15015.
Virgilio, M., Fauvelot, C., Costantini, F., Abbiati, M., & Backeljau, T. (2009). Phylogeography of the common ragworm Hediste diversicolor (Polychaeta: Nereididae) reveals cryptic diversity and multiple colonization events across its distribution. Molecular Ecology, 18(9), 1980–1994. https://doi.org/10.1111/j.1365-294X.2009.04170.x.
Vrijenhoek, R. C. (2009). Cryptic species, phenotypic plasticity, and complex life histories: assessing deep-sea faunal diversity with molecular markers. Deep Sea Research Part II: Topical Studies in Oceanography, 56(19–20), 1713–1723. https://doi.org/10.1016/j.dsr2.2009.05.016.
Webster, H. (1879). Annelida chaetopoda of New Jersey. New York State Museum Annual Report, 32, 101–128.
Wiklund, H., Glover, A. G., Johannessen, P. J., & Dahlgren, T. G. (2009). Cryptic speciation at organic-rich marine habitats: a new bacteriovore annelid from whale-fall and fish farms in the North-East Atlantic. Zoological Journal of the Linnean Society, 155(4), 774–785. https://doi.org/10.1111/j.1096-3642.2008.00469.x.
Wiklund, H., Taylor, J. D., Dahlgren, T. G., Todt, C., Ikebe, C., Rabone, M., & Glover, A. G. (2017). Abyssal fauna of the UK-1 polymetallic nodule exploration area, Clarion-Clipperton Zone, central Pacific Ocean: Mollusca. ZooKeys, 707, 1–46. https://doi.org/10.3897/zookeys.707.13042.
Wiklund, H., Neal, L., Dahlgren, T. G., Drennan, R., Rabone, M., & Glover, A. G. (submitted). Abyssal fauna of polymetallic nodule exploration areas, eastern Clarion-Clipperton Zone, central Pacific Ocean: Annelida: Capitellidae, Opheliidae, Scalibregmatidae and Travisiidae.
Wilson, R. S. (1990). Prionospio and Paraprionospio (Polychaeta: Spionidae) from southern Australia. Memoirs of the Museum of Victoria, 50, 243–274.
Wilson, W. H. (1991). Sexual reproductive modes in polychaetes: classification and diversity. Bulletin of Marine Science, 48, 17.
Won, Y., Young, C. R., Lutz, R. A., & Vrijenhoek, R. C. (2003). Dispersal barriers and isolation among deep-sea mussel populations (Mytilidae: Bathymodiolus) from eastern Pacific hydrothermal vents. Molecular Ecology, 12(1), 169–184. https://doi.org/10.1046/j.1365-294X.2003.01726.x.
Yearsley, J. M., & Sigwart, J. D. (2011). Larval transport modeling of deep-sea invertebrates can aid the search for undiscovered populations. PLoS One, 6(8), e23063. https://doi.org/10.1371/journal.pone.0023063.
Yokoyama, H. (2007). A revision of the genus Paraprionospio Caullery (Polychaeta: Spionidae). Zoological Journal of the Linnean Society, 151(2), 253–284. https://doi.org/10.1111/j.1096-3642.2007.00323.x.
Young, C. (2003). Reproduction, development and life-history traits. In Ecosystems of the world (pp. 381–426). Amsterdam: Elsevier Science.
Young, C. R., Fujio, S., & Vrijenhoek, R. C. (2008). Directional dispersal between mid-ocean ridges: deep-ocean circulation and gene flow in Ridgeia piscesae. Molecular Ecology, 17(7), 1718–1731. https://doi.org/10.1111/j.1365-294X.2008.03609.x.
Zwickl, D. J. (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin: Ph.D thesis, University of Texas at Austin.
Acknowledgments
The authors are grateful to the crew of the RV Sonne RV Melville and RV Thomas G. Thompson for valuable support during the Vema-TRANSIT expedition and the two ABYSSLINE cruises and to many students and colleagues for sample sorting and help on deck. Craig R. Smith (University of Hawaii) and Adrian Glover (Natural History Museum, London) are acknowledged for coordinating the ABYSSLINE project and sampling onboard. Thank you to L. Menot (Ifremer) who participated in the JPI Oceans and MIDAS projects that allowed to sample additional polychaetes from the CCZ. T. Guggolz received a PhD fellowship from the Bauer Foundation (Germany). Special thanks to the GRG, for supporting the first author with valuable comments and new ideas. Marco Neiber is thanked for support and help with statistical tests. Two anonymous reviewers are thanked for valuable comments improving the manuscript.
Funding
Open Access funding provided by Projekt DEAL. The project was undertaken with financial support of the PTJ (German Ministry for Science and Education), grant 03G0227A to A. Brandt and C. Devey and from UK Seabed Resources and the Swedish Research Council FORMAS to H. Wiklund and T.G. Dahlgren. P. Bonifácio received financial support from the JPI Oceans pilot action ‘Ecological Aspects of Deep-Sea Mining’ and from the European Union Seventh Framework Program (FP7/2007–2013) under the MIDAS project, grant agreement no. 603418.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guggolz, T., Meißner, K., Schwentner, M. et al. High diversity and pan-oceanic distribution of deep-sea polychaetes: Prionospio and Aurospio (Annelida: Spionidae) in the Atlantic and Pacific Ocean. Org Divers Evol 20, 171–187 (2020). https://doi.org/10.1007/s13127-020-00430-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-020-00430-7