Abstract
The automatic cell analysis method is capable of segmenting the cells and can detect the number of live/dead cells present in the body. This study proposed a novel non-linear segmentation model (NSM) for the segmentation and quantification of live/dead cells present in the body. This work also reveals the aspects of electromagnetic radiation on the cell body. The bright images of the hippocampal CA3 region of the rat brain under the resolution of 60 × objective are used to analyze the effects called NISSL-stained dataset. The proposed non-linear segmentation model segments the foreground cells from the cell images based on the linear regression analysis. These foreground cells further get discriminated as live/dead cells and quantified using shape descriptors and geometric method, respectively. The proposed segmentation model is showing promising results (accuracy, 82.82%) in comparison with the existing renowned approaches. The counting analysis of live and dead cells using the proposed method is far better than the manual counts. Therefore, the proposed segmentation model and quantifying procedure is an amalgamated method for cell quantification that yields better segmentation results and provides pithy insights into the analysis of neuronal anomalies at a microscopic level.

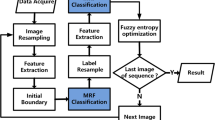
Resultant View of the overall proposed approach



















Similar content being viewed by others
References
World Health Organization (2006) Comprehensive cervical cancer control: a guide to essential practice, World Health Organization Department of Reproductive Health and Research and Department of Chronic Diseases and Health Promotion
Hyland GJ (2000) Physics and biology of mobile telephony. Lancet 356:1833–1836
LG Salford AE, Brun JL, Eberhardt L, Malmgren BR (2003) Persson: Nerve cell damage in mammalian brain after exposure to microwaves from GSM mobile phones. Environ Health Perspect 111(7):881–883
Lin JC (2004) Studies on microwaves in medicine and biology: from snails to humans. Bioelectromagnetics 25 (3):146–159
Belyaev IY, Grigoriev YG (2007) Problems in assessment of risks from exposures to microwaves of mobile communication. Radiat Biol Radioecol 47(6):727–732
Hardell L, Sage C (2008) Biological effects from electromagnetic field exposure and public exposure standards. Biomed Pharmacother 62(2):104–109
Bergmeir C, Silvente MG, Benítez JM (2012) Segmentation of cervical cell nuclei in high-resolution microscopic images: a new algorithm and a web-based software framework. Comput Meth Prog Bio 107(3):497–512
Bamford P, Lovell B (1998) Unsupervised cell nucleus segmentation with active contours. Signal Process 71(2):203–213
Tsai MH, Chan YK, Lin ZZ, Yang-Mao SF, Huang PC (2008) Nucleus and cytoplast contour detector of cervical smear image. Pattern Recogn Lett 29(9):1441–1453
Chang H, Yang Q, Parvin B (2007) Segmentation of heterogeneous blob objects through voting and level set formulation. Pattern Recogn Lett 28(13):1781–1787
Ta VT, Lézoray O, Elmoataz A, Schüpp S (2009) Graph-based tools for microscopic cellular image segmentation. Pattern Recogn 42(6):1113–1125
Pal A, Garain U, Chandra A, Chatterjee R, Senapati S (2018) Psoriasis skin biopsy image segmentation using Deep Convolutional Neural Network. Comput Meth Prog Bio 159:59–69
Moshavash Z, Danyali H, Helfroush MS (2018) An automatic and robust decision support system for accurate acute leukemia diagnosis from blood microscopic images. J Digit Imaging 31:1–16
ko BC, Seo M, Nam J-Y (2009) Microscopic cell nuclei segmentation based on adaptive attention window. J Digit Imaging 22(3):259
Cheng J, Rajapakse JC (2009) Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans Biomed Eng 56(3):741–748
Wu Q, Gan Y, Lin B, Zhang Q, Chang H (2015) An active contour model based on fused texture features for image segmentation. Neurocomputing 151:1133–1141
Molnar C, Jermyn IH, Kato Z, Rahkama V, Östling P, Mikkonen P, Horvath P (2412) Accurate morphology preserving segmentation of overlapping cells based on active contours. Sci Rep 6(3):2016
Ma H, Beiter R, Gaultier A, Acton ST, Lin Z (2018) Olso: automatic cell counting and segmentation for oligodendrocyte progenitor cells. IEEE International Conference on Image Processing
Cochrane D, Orcutt GH (1949) Application of least squares regression to relationships containing auto-correlated error terms. J Am Stat Assoc 44(245):32–61
Zhi X, Jiang S, Zhang W, Wang D, Li Y (2017) Image degradation characteristics and restoration based on regularization for diffractive imaging. Infra-red Phys Technol 86:226–238
Kass RE (1990) Non-linear regression analysis and its applications. J Am Stat Assoc 85(410):594–596
Benzinou A, Hojeij Y, Sibiril Y, Roudot AC (2006) Haematopoietic cell clusters quantification using image analysis. Biomed Signal Process Control 1(4):282–288
Suvarna KS, Layton C, Bancroft JD (2012) Bancroft’s theory and practice of histological techniques e-book. Elsevier Health Sciences
Al-Fahdawi S, Qahwaji R, Al-Waisy AS, Ipson S, Ferdousi M, Malik RA, Brahma A (2018) A fully automated cell segmentation and morphometric parameter system for quantifying corneal endothelial cell morphology. Comput Meth Prog Bio 160: 11–23
Kopanja L, Kovacevic Z, Tadic M, Zuzek MC, Vrecl M, Frangez R (2018) Confocal micrographs: automated segmentation and quantitative shape analysis of neuronal cells treated with ostreolysin A/pleurotolysin B pore-forming complex. Histochem Cell Biol 150:1–10
Albayrak A, Bilgin G (2019) Automatic cell segmentation in histopathological images via two-staged superpixel-based algorithms. Med Biol Eng Comput 57(3):653–665
Kanatani K, Sugaya Y, Kanazawa Y (2016) “Ellipse Fitting.” guide to 3D vision computation. Springer, Cham, pp 11–32
Nishad PM (2013) Various colour spaces and colour space conversion. J Glob Res Comput Sci 4(1):44–48
Wang J, et al. (2019) Automatic cell segmentation and signal detection in fluorescent in situ hybridization. In: Proceedings of 2018 Chinese intelligent systems conference, Springer, Singapore
Acknowledgments
The authors would like to acknowledge that the work is supported by Department of Technology (DoT) and Department of Science and Technology (DST), India. Dataset used in the proposed work is supported and provided by the Manipal University, Department of Life Science Engineering, Karnataka, India.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharma, M., Bhattacharya, M. Discrimination and quantification of live/dead rat brain cells using a non-linear segmentation model. Med Biol Eng Comput 58, 1127–1146 (2020). https://doi.org/10.1007/s11517-020-02135-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-020-02135-7