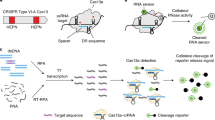
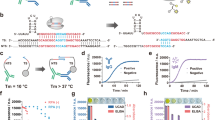
Abstract
In organ transplantation, infection and rejection are major causes of graft loss. They are linked by the net state of immunosuppression. To diagnose and treat these conditions earlier, and to improve long-term patient outcomes, refined strategies for the monitoring of patients after graft transplantation are needed. Here, we show that a fast and inexpensive assay based on CRISPR–Cas13 accurately detects BK polyomavirus DNA and cytomegalovirus DNA from patient-derived blood and urine samples, as well as CXCL9 messenger RNA (a marker of graft rejection) at elevated levels in urine samples from patients experiencing acute kidney transplant rejection. The assay, which we adapted for lateral-flow readout, enables—via simple visualization—the post-transplantation monitoring of common opportunistic viral infections and of graft rejection, and should facilitate point-of-care post-transplantation monitoring.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary information files. The raw and analysed datasets generated during the study are available for research purposes from the corresponding authors on reasonable request, and the availability of raw patient data is subject to approval from the Institutional Review Board.
Code availability
The lateral-flow quantification app code is available at https://github.com/jackievaleri/lateral_flow_quantification_app.
References
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356, 438–442 (2017).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR–Cas13. Science 448, 444–448 (2018).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Chen, J. S. et al. CRISPR–Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Harrington, L. B. et al. Programmed DNA destruction by miniature CRISPR–Cas14 enzymes. Science 362, 839–842 (2018).
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Haijan, R. et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 3, 427–437 (2019).
Wu, Y., Liu, S.-X., Wang, F. & Zeng, M.-S. Room temperature detection of plasma Epstein–Barr virus DNA with CRISPR–Cas13. Clin. Chem. 65, 591–592 (2019).
Lamb, K. E., Lodhi, S. & Meier-Kriesche, H.-U. Long-term renal allograft survival in the United States: a critical reappraisal. Am. J. Transplant. 11, 450–462 (2011).
Tullius, S. G. & Rabb, H. Improving the supply and quality of deceased-donor organs for transplantation. N. Engl. J. Med. 378, 1920–1929 (2018).
Wekerle, T., Segev, D., Lechler, R. & Oberbauer, R. Strategies for long-term preservation of kidney graft function. Lancet 389, 2152–2162 (2017).
Ross, S. A., Novak, Z., Pati, S. & Boppana, S. B. Overview of the diagnosis of cytomegalovirus infection. Infect. Disord. Drug Targets 11, 466–474 (2011).
Randhawa, P. et al. Correlates of quantitative measurement of BK polyomavirus (BKV) DNA with clinical course of BKV infection in renal transplant patients. J. Clin. Microbiol. 42, 1176–1180 (2004).
Fishman, J. A. Infection in organ transplantation. Am. J. Transplant. 17, 856–879 (2017).
Sepkowitz, K. A. Opportunistic infections in patients with and patients without acquired immunodeficiency syndrome. Clin. Infect. Dis. 34, 1098–1107 (2002).
Sundsfjord, A. et al. BK and JC viruses in human immunodeficiency virus type 1-infected persons: prevalence, excretion, viremia, and viral regulatory regions. J. Infect. Dis. 169, 485–490 (1994).
Jackson, J. A. et al. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am. J. Transplant. 11, 2228–2234 (2011).
Hricik, D. E. et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am. J. Transplant. 13, 2634–2644 (2013).
Schaub, S. et al. Urinary CXCL9 and CXCL10 levels correlate with the extent of subclinical tubulitis. Am. J. Transplant. 9, 1347–1353 (2009).
Rabant, M. et al. Early low urinary CXCL9 and CXCL10 might predict immunological quiescence in clinically and histologically stable kidney recipients. Am. J. Transplant. 16, 1868–1881 (2016).
Fishman, J. A. Infection in solid-organ transplant recipients. N. Engl. J. Med. 357, 2601–2614 (2007).
El-Zoghby, Z. M. et al. Identifying specific causes of kidney allograft loss. Am. J. Transplant. 9, 527–535 (2009).
Henderson, L. K., Nankivell, B. J. & Chapman, J. R. Surveillance protocol kidney transplant biopsies: their evolving role in clinical practice. Am. J. Transplant. 11, 1570–1575 (2011).
Bloom, R. D. et al. Cell-free DNA and active rejection in kidney allografts. J. Am. Soc. Nephrol. 28, 2221–2232 (2017).
Sigdel, T. K. et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J. Clin. Med. 8, E19 (2018).
Hricik, D. E. et al. Adverse outcomes of tacrolimus withdrawal in immune–quiescent kidney transplant recipients. J. Am. Soc. Nephrol. 26, 3114–3122 (2015).
Curtis, K. A. et al. Isothermal amplification using a chemical heating device for point-of-care detection of HIV-1. PLoS ONE 7, e31432 (2012).
Ahn, M.-H., Baek, S.-K., Min, J. & Park, J.-H. A portable electromagnetic induction heating device for point-of-care diagnostics. BioChip J. 10, 208–214 (2016).
Parajuli, S. et al. Donor-specific antibodies in the absence of rejection are not a risk factor for allograft failure. Kidney Int. Rep. 4, 1057–1065 (2019).
Schinstock, C. A. et al. The value of protocol biopsies to identify patients with de novo donor-specific antibody at high risk for allograft loss. Am. J. Transplant. 17, 1574–1584 (2017).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101 (2008).
Acknowledgements
We thank R. Zaffini from viral microbiology (BWH) for helpful discussion about BKV testing and qPCR; the Crimson Core at BWH for providing samples from patients with CMV and BKV; and H. de Puig, N. Angenent-Mari, A. Dy and X. Tan for helpful discussions. M.M.K. was supported by the German Academy of Sciences, Leopoldina (LPDS 2018-01), the Clinician Scientist Program Berta–Ottenstein of the Faculty of Medicine, University of Freiburg, Germany and the Emmy Noether Programme (KA5060/1-1). M.A.A. was supported by a National Science Foundation graduate research fellowship (award no. 1122374). V.K. was supported by the DFG (grant no. 403877094). J.J.C. was supported by MIT’s Center for Microbiome Informatics and Therapeutics, the Paul G. Allen Frontiers Group and the Wyss Institute.
Author information
Authors and Affiliations
Contributions
M.M.K., L.V.R. and J.J.C. designed the study. M.M.K., M.A.A., A.C.H., I.L. and R.G. performed experiments. J.A.V. and M.A.A. programmed the smartphone app, and V.K. contributed to the design and analysis of experiments. L.V.R. and E.A. provided clinical samples. F.M.M. and J.A. advised on the collection and testing of clinical specimens. All authors contributed to the writing of the manuscript and interpretation of data.
Corresponding authors
Ethics declarations
Competing interests
A patent application related to this work is pending. J.J.C. is co-founder and director of Sherlock Biosciences.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures and tables.
Supplementary Video 1
A smartphone-based software application for the quantification of band intensities.
Rights and permissions
About this article
Cite this article
Kaminski, M.M., Alcantar, M.A., Lape, I.T. et al. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat Biomed Eng 4, 601–609 (2020). https://doi.org/10.1038/s41551-020-0546-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-020-0546-5
This article is cited by
-
Recent Advances in Electrochemical and Nanophotonic Biosensors for SARS-CoV-2 Detection
BioChip Journal (2024)
-
Potential of CRISPR/Cas system as emerging tools in the detection of viral hepatitis infection
Virology Journal (2023)
-
CRISPR-Cas-amplified urinary biomarkers for multiplexed and portable cancer diagnostics
Nature Nanotechnology (2023)
-
Four thermostatic steps: A novel CRISPR-Cas12-based system for the rapid at-home detection of respiratory pathogens
Applied Microbiology and Biotechnology (2023)
-
Rejection markers in kidney transplantation: do new technologies help children?
Pediatric Nephrology (2023)