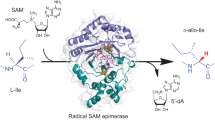
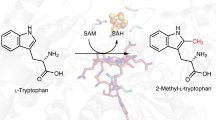
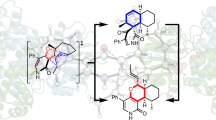
Abstract
Radical S-adenosyl-l-methionine (SAM) enzymes belong to a family of catalysts whose number of annotated sequences is still growing. Upon the one-electron reduction of a [Fe4S4] cluster, they can cleave SAM to produce a highly reactive 5′-deoxyadenosyl radical species. This radical species in turn triggers a wide variety of radical-based reactions on substrates ranging from small organic molecules to proteins, DNA or RNA. The challenging reactions they catalyse makes them very promising catalysts for diverse biotechnological applications. However, the high-energy intermediates involved require fine control of the chemistry by the protein matrix. Understanding their control mechanism is a prerequisite for a broader use of these enzymes as synthetic tools. Here I review some of the latest developments in the field, focusing on the structure–function relationship of a few examples for which three-dimensional structures, in vitro and spectroscopic data, as well as theoretical calculations, are available to better describe the close interaction between the chemistry performed and the tight control of the protein matrix.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Togo, H. Advanced Free Radical Reactions for Organic Synthesis (Elsevier Science, 2004).
Frey, P. A., Hegeman, A. D. & Ruzicka, F. J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 (2008).
Broderick, J. B., Duffus, B. R., Duschene, K. S. & Shepard, E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014).
Bridwell-Rabb, J., Grell, T. A. J. & Drennan, C. L. A Rich man, poor man story of S-adenosylmethionine and cobalamin revisited. Annu. Rev. Biochem. 87, 555–584 (2018).
Stubbe, J. Binding site revealed of nature’s most beautiful cofactor. Science 266, 1663–1664 (1994).
Martens, J. H., Barg, H., Warren, M. J. & Jahn, D. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58, 275–285 (2002).
Frey, P. A. Lysine 2,3-aminomutase: is adenosylmethionine a poor man’s adenosylcobalamin? FASEB J. 7, 662–670 (1993).
Sofia, H. J., Chen, G., Hetzler, B. G., Reyes-Spindola, J. F. & Miller, N. E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 (2001). This seminal paper describes radical SAM enzymes as a superfamily.
Akiva, E. et al. The Structure–Function Linkage Database. Nucleic Acids Res. 42, D521–D530 (2014).
Begley, T. P., Xi, J., Kinsland, C., Taylor, S. & McLafferty, F. The enzymology of sulfur activation during thiamin and biotin biosynthesis. Curr. Opin. Chem. Biol. 3, 623–629 (1999).
Miller, J. R. et al. Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39, 15166–15178 (2000).
Berteau, O., Guillot, A., Benjdia, A. & Rabot, S. A new type of bacterial sulfatase reveals a novel maturation pathway in prokaryotes. J. Biol. Chem. 281, 22464–22470 (2006).
Anton, B. P. et al. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc. Natl Acad. Sci. USA 105, 1826–1831 (2008).
Yan, F. et al. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J. Am. Chem. Soc. 132, 3953–3964 (2010).
Sun, X. et al. Generation of the glycyl radical of the anaerobic Escherichia coli ribonucleotide reductase requires a specific activating enzyme. J. Biol. Chem. 270, 2443–2446 (1995).
Rebeil, R. et al. Spore photoproduct lyase from Bacillus subtilis spores is a novel iron-sulfur DNA repair enzyme which shares features with proteins such as class III anaerobic ribonucleotide reductases and pyruvate-formate lyases. J. Bacteriol. 180, 4879–4885 (1998).
Yokoyama, K. & Lilla, E. A. C-C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products. Nat. Prod. Rep. 35, 660–694 (2018).
Ruszczycky, M. W., Ogasawara, Y. & Liu, H.-W. Radical SAM enzymes in the biosynthesis of sugar-containing natural products. Biochim. Biophys. Acta 1824, 1231–1244 (2012).
Benjdia, A., Balty, C. & Berteau, O. Radical SAM enzymes in the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs). Front. Chem. 5, 87 (2017).
Jaeger, C. M. & Croft, A. K. Anaerobic radical enzymes for biotechnology. Chembioeng. Rev. 5, 143–162 (2018).
Broderick, W. E., Hoffman, B. M. & Broderick, J. B. Mechanism of radical initiation in the radical S-adenosyl-l-methionine superfamily. Acc. Chem. Res. 51, 2611–2619 (2018).
Vey, J. L. & Drennan, C. L. Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 (2011).
McGlynn, S. E. et al. Identification and characterization of a novel member of the radical AdoMet enzyme superfamily and implications for the biosynthesis of the Hmd hydrogenase active site cofactor. J. Bacteriol. 192, 595–598 (2010).
Kim, H. J., LeVieux, J., Yeh, Y.-C. & Liu, H. C3-deoxygenation of paromamine catalyzed by a radical S-adenosylmethionine enzyme: characterization of the enzyme AprD4 and its reductase partner AprD3. Angew. Chem. Int. Ed. 55, 3724–3728 (2016).
Dowling, D. P. et al. Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat. Chem. Biol. 10, 106–112 (2014).
Bridwell-Rabb, J., Zhong, A., Sun, H. G., Drennan, C. L. & Liu, H.-W. A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis. Nature 544, 322–326 (2017).
Fenwick, M. K. et al. Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase. Nat. Commun. 6, 6480 (2015).
Walsby, C. J., Ortillo, D., Broderick, W. E., Broderick, J. B. & Hoffman, B. M. An anchoring role for FeS clusters: chelation of the amino acid moiety of. J. Am. Chem. Soc. 124, 11270–11271 (2002).
Walsby, C. J. et al. Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe–4S]+ cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124, 3143–3151 (2002).
Layer, G., Moser, J., Heinz, D. W., Jahn, D. & Schubert, W.-D. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 22, 6214–6224 (2003).
Berkovitch, F., Nicolet, Y., Wan, J. T., Jarrett, J. T. & Drennan, C. L. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 (2004).
Frey, P. A. Travels with carbon-centered radicals. 5′-deoxyadenosine and 5′-deoxyadenosine-5′-yl in radical enzymology. Acc. Chem. Res. 47, 540–549 (2014).
Wang, S. C. & Frey, P. A. Binding energy in the one-electron reductive cleavage of S-adenosylmethionine in lysine 2,3-aminomutase, a radical SAM enzyme. Biochemistry 46, 12889–12895 (2007).
Canfield, J. M. & Warncke, K. Geometry of reactant centers in the Coii-substrate radical pair state of coenzyme B12-dependent ethanolamine deaminase determined by using orientation-selection-ESEEM spectroscopy. J. Phys. Chem. B 106, 8831–8841 (2002).
Maity, A. N. et al. Evidence for conformational movement and radical mechanism in the reaction of 4-thia-l-lysine with lysine 5,6-aminomutase. J. Phys. Chem. B 113, 12161–12163 (2009).
LoBrutto, R. et al. 5′-Deoxyadenosine contacts the substrate radical intermediate in the active site of ethanolamine ammonia-lyase: 2H and 13C electron nuclear double resonance studies. Biochemistry 40, 9–14 (2001).
Magnusson, O. T., Reed, G. H. & Frey, P. A. Characterization of an allylic analogue of the 5′-deoxyadenosyl radical: an intermediate in the reaction of lysine 2,3-aminomutase. Biochemistry 40, 7773–7782 (2001).
Rohac, R. et al. Carbon–sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE. Nat. Chem. 8, 491–500 (2016). Report on a radical-based reaction triggered and observed directly in crystallo.
Horitani, M. et al. Why nature uses radical SAM enzymes so widely: electron nuclear double resonance studies of lysine 2,3-aminomutase show the 5′-dAdo* ‘free radical’ is never free. J. Am. Chem. Soc. 137, 7111–7121 (2015).
Miller, S. A. & Bandarian, V. Analysis of electrochemical properties of S-adenosyl-l-methionine and implications for its role in radical SAM enzymes. J. Am. Chem. Soc. 141, 11019–11026 (2019).
Goldman, P. J., Grove, T. L., Booker, S. J. & Drennan, C. L. X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proc. Natl Acad. Sci. USA 110, 15949–15954 (2013).
Nicolet, Y., Zeppieri, L., Amara, P. & Fontecilla-Camps, J. C. Crystal structure of tryptophan lyase (NosL): evidence for radical formation at the amino group of tryptophan. Angew. Chem. Int. Ed. 53, 11840–11844 (2014).
Benjdia, A., Heil, K., Barends, T. R. M., Carell, T. & Schlichting, I. Structural insights into recognition and repair of UV-DNA damage by spore photoproduct lyase, a radical SAM enzyme. Nucleic Acids Res. 40, 9308–9318 (2012).
Lepore, B. W., Ruzicka, F. J., Frey, P. A. & Ringe, D. The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. Proc. Natl Acad. Sci. USA 102, 13819–13824 (2005).
Goldman, P. J. et al. X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl Acad. Sci. USA 110, 8519–8524 (2013).
Liu, W.-Q. et al. 1,2-Diol dehydration by the radical SAM enzyme AprD4: a matter of proton circulation and substrate flexibility. J. Am. Chem. Soc. 140, 1365–1371 (2018).
Nicolet, Y., Amara, P., Mouesca, J.-M. & Fontecilla-Camps, J. C. Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins. Proc. Natl Acad. Sci. USA 106, 14867–14871 (2009).
Cosper, N. J., Booker, S. J., Ruzicka, F., Frey, P. A. & Scott, R. A. Direct FeS cluster involvement in generation of a radical in lysine 2,3-aminomutase. Biochemistry 39, 15668–15673 (2000).
Horitani, M. et al. Radical SAM catalysis via an organometallic intermediate with an Fe-[5′-C]-deoxyadenosyl bond. Science 352, 822–825 (2016).
Byer, A. S. et al. Paradigm shift for radical S-adenosyl-l-methionine reactions: the organometallic intermediate omega is central to catalysis. J. Am. Chem. Soc. 140, 8634–8638 (2018).
Yang, H. et al. The elusive 5′-deoxyadenosyl radical: captured and characterized by electron paramagnetic resonance and electron nuclear double resonance spectroscopies. J. Am. Chem. Soc. 141, 12139–12146 (2019). Observation and characterization of the long-sought-after 5′-deoxyadenosyl radical species.
Yang, H. et al. Photoinduced electron transfer in a radical SAM enzyme generates an S-adenosylmethionine derived methyl radical. J. Am. Chem. Soc. 141, 16117–16124 (2019).
Vey, J. L. et al. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc. Natl Acad. Sci. USA 105, 16137–16141 (2008).
Nicolet, Y. et al. Crystal structure of HydG from Carboxydothermus hydrogenoformans: a trifunctional [FeFe]-hydrogenase maturase. Chembiochem 16, 397–402 (2015).
Sayler, R. I. et al. Trapping and electron paramagnetic resonance characterization of the 5′dAdo• radical in a radical S-adenosyl methionine enzyme reaction with a non-native substrate. ACS Cent. Sci. 5, 1777–1785 (2019).
Toraya, T. Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem. Rev. 103, 2095–2127 (2003).
Yokoyama, K., Numakura, M., Kudo, F., Ohmori, D. & Eguchi, T. Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. J. Am. Chem. Soc. 129, 15147–15155 (2007).
Szu, P., He, X., Zhao, L. & Liu, H. Biosynthesis of TDP-d-desosamine: identification of a strategy for C4 deoxygenation. Angew. Chem. Int. Ed. 44, 6742–6746 (2005).
Lv, M. et al. Characterization of a C3 deoxygenation pathway reveals a key branch point in aminoglycoside biosynthesis. J. Am. Chem. Soc. 138, 6427–6435 (2016).
Liu, W.-Q. et al. 1,2-Diol dehydration by the radical SAM enzyme AprD4: a matter of proton circulation and substrate flexibility. J. Am. Chem. Soc. 140, 1365–1371 (2018).
Grove, T. L., Ahlum, J. H., Sharma, P., Krebs, C. & Booker, S. J. A consensus mechanism for radical SAM-dependent dehydrogenation? BtrN contains two [4Fe–4S] clusters. Biochemistry 49, 3783–3785 (2010).
Grell, T. A. J., Goldman, P. J. & Drennan, C. L. SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J. Biol. Chem. 290, 3964–3971 (2015).
Yokoyama, K., Ohmori, D., Kudo, F. & Eguchi, T. Mechanistic study on the reaction of a radical SAM dehydrogenase BtrN by electron paramagnetic resonance spectroscopy. Biochemistry 47, 8950–8960 (2008).
Hayon, E. & Simic, M. Acid-base properties of free-radicals in solution. Acc. Chem. Res. 7, 114–121 (1974).
Maiocco, S. J., Grove, T. L., Booker, S. J. & Elliott, S. J. Electrochemical resolution of the [4Fe–4S] centers of the AdoMet radical enzyme BtrN: evidence of proton coupling and an unusual, low-potential auxiliary cluster. J. Am. Chem. Soc. 137, 8664–8667 (2015).
Ruszczycky, M. W. & Liu, H. Mechanistic enzymology of the radical SAM enzyme DesII. Isr. J. Chem. 55, 315–324 (2015).
Benitez-Paez, A., Villarroya, M. & Armengod, M.-E. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA 18, 1783–1795 (2012).
Yan, F. & Fujimori, D. G. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc. Natl Acad. Sci. USA 108, 3930–3934 (2011).
Grove, T. L. et al. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science 332, 604–607 (2011).
Grove, T. L., Radle, M. I., Krebs, C. & Booker, S. J. Cfr and RlmN contain a single [4Fe–4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: methyl transfer by SN2 displacement and radical generation. J. Am. Chem. Soc. 133, 19586–19589 (2011).
Boal, A. K. et al. Structural basis for methyl transfer by a radical SAM enzyme. Science 332, 1089–1092 (2011).
Wu, W., Lieder, K. W., Reed, G. H. & Frey, P. A. Observation of a second substrate radical intermediate in the reaction of lysine 2,3-aminomutase: a radical centered on the beta-carbon of the alternative substrate, 4-thia-l-lysine. Biochemistry 34, 10532–10537 (1995).
McCusker, K. P. et al. Covalent intermediate in the catalytic mechanism of the radical S-adenosyl-l-methionine methyl synthase RlmN trapped by mutagenesis. J. Am. Chem. Soc. 134, 18074–18081 (2012).
Silakov, A. et al. Characterization of a cross-linked protein-nucleic acid substrate radical in the reaction catalyzed by RlmN. J. Am. Chem. Soc. 136, 8221–8228 (2014).
Grove, T. L. et al. A substrate radical intermediate in catalysis by the antibiotic resistance protein Cfr. Nat. Chem. Biol. 9, 422–427 (2013).
Schwalm, E. L., Grove, T. L., Booker, S. J. & Boal, A. K. Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA. Science 352, 309–312 (2016). An elegant capture of an intermediate between a radical SAM and its tRNA substrate.
Zhao, C., Dong, L. & Liu, Y. A QM/MM study of the catalytic mechanism of SAM methyltransferase RlmN from Escherichia coli. Proteins 85, 1967–1974 (2017).
McCarty, R. M., Somogyi, A. & Bandarian, V. Escherichia coli QueD is a 6-carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry 48, 2301–2303 (2009).
McCarty, R. M., Somogyi, A., Lin, G., Jacobsen, N. E. & Bandarian, V. The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of PreQ(0) from guanosine 5′-triphosphate in four steps. Biochemistry 48, 3847–3852 (2009).
McCarty, R. M., Krebs, C. & Bandarian, V. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines. Biochemistry 52, 188–198 (2013).
Zhu, W. & Liu, Y. Ring contraction catalyzed by the metal-dependent radical SAM enzyme: 7-carboxy-7-deazaguanine synthase from B. multivorans. Theoretical insights into the reaction mechanism and the influence of metal ions. ACS Catal. 5, 3953–3965 (2015).
Jaeger, C. M. & Croft, A. K. Radical reaction control in the AdoMet radical enzyme CDG synthase (QueE): consolidate, destabilize, accelerate. Chemistry 23, 953–962 (2017).
Zhang, Q. et al. Radical-mediated enzymatic carbon chain fragmentation-recombination. Nat. Chem. Biol. 7, 154–160 (2011).
Yu, Y. et al. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem. Biol. 4, 855–864 (2009).
Nicolet, Y. & Drennan, C. L. AdoMet radical proteins—from structure to evolution—alignment of divergent protein sequences reveals strong secondary structure element conservation. Nucleic Acids Res. 32, 4015–4025 (2004).
Quitterer, F., List, A., Eisenreich, W., Bacher, A. & Groll, M. Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis. Angew. Chem. Int. Ed. 51, 1339–1342 (2012).
Nicolet, Y. et al. X-ray structure of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima. J. Biol. Chem. 283, 18861–18872 (2008).
Bhandari, D. M., Xu, H., Nicolet, Y., Fontecilla-Camps, J. C. & Begley, T. P. Tryptophan lyase (NosL): mechanistic insights from substrate analogues and mutagenesis. Biochemistry 54, 4767–4769 (2015).
Ji, X., Li, Y., Ding, W. & Zhang, Q. Substrate-tuned catalysis of the radical S-adenosyl-l-methionine enzyme NosL involved in nosiheptide biosynthesis. Angew. Chem. Int. Ed. 54, 9021–9024 (2015).
Bhandari, D. M., Fedoseyenko, D. & Begley, T. P. Tryptophan lyase (NosL): a cornucopia of 5′-deoxyadenosyl radical mediated transformations. J. Am. Chem. Soc. 138, 16184–16187 (2016).
Ding, W., Ji, X., Li, Y. & Zhang, Q. Catalytic promiscuity of the radical S-adenosyl-l-methionine enzyme NosL. Front. Chem. 4, 27 (2016).
Ji, X. et al. Expanding radical SAM chemistry by using radical addition reactions and SAM analogues. Angew. Chem. Int. Ed. 55, 11845–11848 (2016).
Kuchenreuther, J. M. et al. A radical intermediate in tyrosine scission to the CO and CN– ligands of FeFe hydrogenase. Science 342, 472–475 (2013).
Kriek, M., Martins, F., Challand, M. R., Croft, A. & Roach, P. L. Thiamine biosynthesis in Escherichia coli: identification of the intermediate and by-product derived from tyrosine. Angew. Chem. Int. Ed. 46, 9223–9226 (2007).
Sicoli, G. et al. Fine-tuning of a radical-based reaction by radical S-adenosyl-l-methionine tryptophan lyase. Science 351, 1320–1323 (2016). Trapping of an unexpected radical intermediate and its structural determination by EPR.
Amara, P. et al. Radical S-adenosyl-l-methionine tryptophan lyase (NosL): how the protein controls the carboxyl radical •CO2 − migration. J. Am. Chem. Soc. 140, 16661–16668 (2018).
Bhandari, D. M., Fedoseyenko, D. & Begley, T. P. Mechanistic studies on tryptophan lyase (NosL): identification of cyanide as a reaction product. J. Am. Chem. Soc. 140, 542–545 (2018).
Yokoyama, K., Numakura, M., Kudo, F., Ohmori, D. & Eguchi, T. Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. J. Am. Chem. Soc. 129, 15147–15155 (2007).
Szu, P.-H., Ruszczycky, M. W., Choi, S., Yan, F. & Liu, H. Characterization and mechanistic studies of DesII: a radical S-adenosyl-l-methionine enzyme involved in the biosynthesis of TDP-d-desosamine. J. Am. Chem. Soc. 131, 14030–14042 (2009).
Chang, C. H., Ballinger, M. D., Reed, G. H. & Frey, P. A. Lysine 2,3-aminomutase: rapid mix–freeze–quench electron paramagnetic resonance studies establishing the kinetic competence of a substrate-based radical intermediate. Biochemistry 35, 11081–11084 (1996).
Bruender, N. A., Young, A. P. & Bandarian, V. Chemical and biological reduction of the radical SAM enzyme. Biochemistry 54, 2903–2910 (2015).
Arcinas, A. J., Maiocco, S. J., Elliott, S. J., Silakov, A. & Booker, S. J. Ferredoxins as interchangeable redox components in support of MiaB, a radical S-adenosylmethionine methylthiotransferase. Protein Sci. 28, 267–282 (2019).
Grove, T. L. et al. Further characterization of Cys-type and Ser-type anaerobic sulfatase maturating enzymes suggests a commonality in the mechanism of catalysis. Biochemistry 52, 2874–2887 (2013).
Walker, L. M., Kincannon, W. M., Bandarian, V. & Elliott, S. J. Deconvoluting the reduction potentials for the three [4Fe–4S] clusters in an AdoMet radical SCIFF maturase. Biochemistry 57, 6050–6053 (2018).
Ayikpoe, R. et al. Spectroscopic and electrochemical characterization of the mycofactocin biosynthetic protein, MftC, provides insight into its redox flipping mechanism. Biochemistry 58, 940–950 (2019).
Ruszczycky, M. W., Zhong, A. & Liu, H.-W. Following the electrons: peculiarities in the catalytic cycles of radical SAM enzymes. Nat. Prod. Rep. 35, 615–621 (2018). A nice review highlighting the role of the redox partner in the efficient catalysis performed by radical SAM enzymes.
Ji, X., Li, Y., Jia, Y., Ding, W. & Zhang, Q. Mechanistic Insights into the radical S-adenosyl-l-methionine enzyme NosL from a substrate analogue and the shunt products. Angew. Chem. Int. Ed. 55, 3334–3337 (2016).
Bhandari, D. M., Fedoseyenko, D. & Begley, T. P. Mechanistic studies on the radical SAM enzyme tryptophan lyase (NosL). Methods Enzymol. 606, 155–178 (2018).
Liu, W., Zhang, Q. & Chen, S. Novel fluoronosiheptide and preparation method and application thereof. Chinese patent CN102453077A (2013).
Wilcoxen, J., Bruender, N. A., Bandarian, V. & Britt, R. D. A radical intermediate in Bacillus subtilis QueE during turnover with the substrate analogue 6-carboxypterin. J. Am. Chem. Soc. 140, 1753–1759 (2018).
Bruender, N. A. et al. 7-Carboxy-7-deazaguanine synthase: a radical S-adenosyl-l-methionine enzyme with polar tendencies. J. Am. Chem. Soc. 139, 1912–1920 (2017).
Qianzhu, H. et al. Reactivity of the nitrogen-centered tryptophanyl radical in the catalysis by the radical SAM enzyme NosL. Chem. Commun. 53, 344–347 (2016).
Benjdia, A., Heil, K., Winkler, A., Carell, T. & Schlichting, I. Rescuing DNA repair activity by rewiring the H-atom transfer pathway in the radical SAM enzyme, spore photoproduct lyase. Chem. Commun. 50, 14201–14204 (2014).
Kruger, T. et al. Conversion of serine-type aldehyde tags by the radical SAM protein AtsB from Methanosarcina mazei. Chembiochem 20, 2074–2078 (2019).
Suess, C. J., Martins, F. L., Croft, A. K. & Jager, C. M. Radical stabilization energies for enzyme engineering: tackling the substrate scope of the radical enzyme QueE. J. Chem. Inf. Model. 59, 5111–5125 (2019).
Arnison, P. G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
Freeman, M. F., Helf, M. J., Bhushan, A., Morinaka, B. I. & Piel, J. Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium. Nat. Chem. 9, 387–395 (2017).
Morinaka, B. I. et al. Radical S-adenosyl methionine epimerases: regioselective introduction of diverse d-amino acid patterns into peptide natural products. Angew. Chem. Int. Ed. 53, 8503–8507 (2014).
Morinaka, B. I. et al. Natural noncanonical protein splicing yields products with diverse beta-amino acid residues. Science 359, 779–782 (2018). An astonishing peptide modification performed by a radical SAM enzyme.
Nordlund, P. & Reichard, P. Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 (2006).
Kampmeier, J. A. Regioselectivity in the homolytic cleavage of S-adenosylmethionine. Biochemistry 49, 10770–10772 (2010).
Dong, M. et al. Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis. Science 359, 1247–1250 (2018).
Acknowledgements
I thank the Commissariat à l’Energie Atomique et aux Energies Alternatives for institutional support. Part of the work presented was supported by the Radis-Bio contract from the CEA/DRF-Impulsion program and the Agence Nationale pour la Recherche (ANR-16-CE29-0019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nicolet, Y. Structure–function relationships of radical SAM enzymes. Nat Catal 3, 337–350 (2020). https://doi.org/10.1038/s41929-020-0448-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-020-0448-7
This article is cited by
-
L-tyrosine-bound ThiH structure reveals C–C bond break differences within radical SAM aromatic amino acid lyases
Nature Communications (2022)
-
Iron–sulfur clusters as inhibitors and catalysts of viral replication
Nature Chemistry (2022)
-
QM/MM Study of the Mechanism of the Noncanonical S-Cγ Bond Scission in S-Adenosylmethionine Catalyzed by the CmnDph2 Radical Enzyme
Topics in Catalysis (2022)
-
Computational identification of a systemic antibiotic for Gram-negative bacteria
Nature Microbiology (2022)