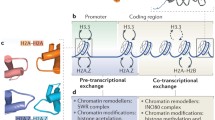
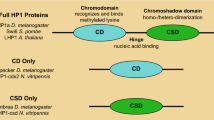
Abstract
The nucleosome is a small unit of chromatin, which is dynamic in eukaryotes. Chromatin conformation and post-translational modifications affect nucleosome dynamics under certain conditions, playing an important role in the epigenetic regulation of transcription, replication and reprogramming. The Snf2 remodeling family is one of the crucial remodeling complexes that tightly regulate chromatin structure and affect nucleosome dynamics. This family alters nucleosome positioning, exchanges histone variants, and assembles and disassembles nucleosomes at certain locations. Moreover, the Snf2 family, in conjunction with other co-factors, regulates gene expression in Saccharomyces cerevisiae. Here we first review recent findings on the Snf2 family remodeling complexes and then use some examples to illustrate the cooperation between different members of Snf2 family, and the cooperation between Snf2 family and other co-factors in gene regulation especially during transcription initiation.




Similar content being viewed by others
References
Ansari SA, Paul E, Sommer S, Lieleg C, He Q, Daly AZ, Rode KA, Barber WT, Ellis LC, LaPorta E, Orzechowski AM, Taylor E, Reeb T, Wong J, Korber P, Morse RH (2014) Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast. J Biol Chem 289:14981–14995
Bao Y, Shen X (2007) INO80 subfamily of chromatin remodeling complexes. Mutat Res 618:18–29
Becker PB, Workman JL (2013) Nucleosome remodeling and epigenetics. Cold Spring Harbor Perspect Biol 5:a017905
Biggar SR, Crabtree GR (1999) Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J 18:2254–2264
Biswas D, Dutta-Biswas R, Stillman DJ (2007) Chd1 and yFACT act in opposition in regulating transcription. Mol Cell Biol 27:6279–6287
Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee TH, Bartholomew B (2017) INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun 8:15616
Cairns BR (2009) The logic of chromatin architecture and remodelling at promoters. Nature 461:193–198
Carlson M, Osmond BC, Neigeborn L, Botstein D (1984) A suppressor of snf1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics 107:19–32
Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304
Clapier CR, Iwasa J, Cairns BR, Peterson CL (2017) Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Bio 18:407–422
Clapier CR, Kasten MM, Parnell TJ, Viswanathan R, Szerlong H, Sirinakis G, Zhang Y, Cairns BR (2016) Regulation of DNA translocation efficiency within the chromatin remodeler RSC/Sth1 potentiates nucleosome sliding and ejection. Mol Cell 62:453–461
Cosgrove MS, Wolberger C (2005) How does the histone code work? Biochem Cell Biol 83:468–476
Daignan-Fornier B, Fink GR (1992) Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA 89:6746–6750
Doyon Y, Cote J (2004) The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev 14:147–154
Farnung L, Vos SM, Wigge C, Cramer P (2017) Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550:539–542
Flaus A, Martin DM, Barton GJ, Owen-Hughes T (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34:2887–2905
Ford J, Odeyale O, Eskandar A, Kouba N, Shen CH (2007) A SWI/SNF- and INO80-dependent nucleosome movement at the INO1 promoter. Biochem Biophys Res Commun 361:974–979
Ford J, Odeyale O, Shen CH (2008) Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation. Biochem Biophys Res Commun 373:602–606
Gerhold CB, Gasser SM (2014) INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol 24:619–631
Gerhold CB, Hauer MH, Gasser SM (2015) INO80-C and SWR-C: guardians of the genome. J Mol Biol 427:637–651
Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423–433
Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W (1999) Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J 18:6407–6414
Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW (2003) Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell 12:449–460
Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L (2005) Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3:e384
Hall JA, Georgel PT (2007) CHD proteins: a diverse family with strong ties. Biochem Cell Biol 85:463–476
Hartley PD, Madhani HD (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137:445–458
Hassan AH, Neely KE, Workman JL (2001) Histone acetyltransferase complexes stabilize Swi/Snf binding to promoter nucleosomes. Cell 104:817–827
Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369–379
Henikoff S, Smith MM (2015) Histone variants and epigenetics. CSH Perspect Biol 7:a019364
Hota SK, Bhardwaj SK, Deindl S, Lin YC, Zhuang XW, Bartholomew B (2013) Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat Struct Mol Biol 20:222–229
Ioshikhes IP, Albert I, Zanton SJ, Pugh BF (2006) Nucleosome positions predicted through comparative genomics. Nat Genet 38:1210–1215
Jiang CZ, Pugh BF (2009) Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10:161–172
Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B (2004) Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J 23:2092–2104
Kasten MM, Clapier CR, Cairns BR (2011) SnapShot: Chromatin remodeling: SWI/SNF. Cell 144:310.e1. https://doi.org/10.1016/j.cell.2011.01.007
Kent NA, Karabetsou N, Politis PK, Mellor J (2001) In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev 15:619–626
Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ (2006) Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol Cell Biol 26:8607–8622
Kingston RE, Tamkun JW (2014) Transcriptional regulation by trithorax-group proteins. CSH Perspect Biol 6:a019349
Koehler RN, Rachfall N, Rolfes RJ (2007) Activation of the ADE genes requires the chromatin remodeling complexes SAGA and SWI/SNF. Eukaryot Cell 6:1474–1485
Kornberg RD (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184:868–871
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P (2016) Genomic nucleosome organization reconstituted with pure proteins. Cell 167:709–721
Lans H, Marteijn JA, Vermeulen W (2012) ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet Chromatin 5:4
Lariviere L, Seizl M, Cramer P (2012) A structural perspective on mediator function. Curr Opin Cell Biol 24:305–313
Lee Y, Park D, Iyer VR (2017) The ATP-dependent chromatin remodeler Chd1 is recruited by transcription elongation factors and maintains H3K4me3/H3K36me3 domains at actively transcribed and spliced genes. Nucleic Acids Res 45:7180–7190
Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128:707–719
Liu X, Li M, Xia X, Li X, Chen Z (2017) Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544:440–445
Lorch Y, Kornberg RD (2015) Chromatin-remodeling and the initiation of transcription. Q Rev Biophys 48:465–470
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260
Lusser A, Urwin DL, Kadonaga JT (2005) Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12:160–166
Malik HS, Henikoff S (2003) Phylogenomics of the nucleosome. Nat Struct Biol 10:882–891
Marfella CG, Imbalzano AN (2007) The Chd family of chromatin remodelers. Mutat Res 618:30–40
Martens JA, Winston F (2002) Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev 16:2231–2236
Martens JA, Wu PY, Winston F (2005) Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev 19:2695–2704
Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O (2015) SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol 10:145–171
McConnell AD, Gelbart ME, Tsukiyama T (2004) Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol Cell Biol 24:2605–2613
Mellor J, Morillon A (2004) ISWI complexes in Saccharomyces cerevisiae. Bioch Biophys Acta 1677:100–112
Menezes RA, Amaral C, Delaunay A, Toledano M, Rodrigues-Pousada C (2004) Yap8p activation in Saccharomyces cerevisiae under arsenic conditions. FEBS Lett 566:141–146
Menezes RA, Pimentel C, Silva AR, Amaral C, Merhej J, Devaux F, Rodrigues-Pousada C (2017) Mediator, SWI/SNF and SAGA complexes regulate Yap8-dependent transcriptional activation of ACR2 in response to arsenate. Bioch Biophys Acta Gene Regul Mech 1860:472–481
Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343–348
Mohanty B, Helder S, Silva APG, Mackay JP, Ryan DP (2016) The chromatin remodelling protein CHD1 contains a previously unrecognised C-terminal helical domain. J Mol Biol 428:4298–4314
Morillon A, Karabetsou N, O'Sullivan J, Kent N, Proudfoot N, Mellor J (2003) Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115:425–435
Morrison AJ (2017) Genome maintenance functions of the INO80 chromatin remodeller. Philos Trans R Soc B 372:1731
Mueller-Planitz F, Klinker H, Ludwigsen J, Becker PB (2013) The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat Struct Mol Biol 20:82–U107
Nodelman IM, Horvath KC, Levendosky RF, Winger J, Ren R, Patel A, Li M, Wang MD, Roberts E, Bowman GD (2016) The Chd1 chromatin remodeler can sense both entry and exit sides of the nucleosome. Nucleic Acids Res 44:7580–7591
Ocampo J, Chereji RV, Eriksson PR, Clark DJ (2016) The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res 44:4625–4635
Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL (2011) Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144:200–213
Parnell EJ, Stillman DJ (2019) Multiple negative regulators restrict recruitment of the SWI/SNF chromatin remodeler to the HO promoter in Saccharomyces cerevisiae. Genetics 212:1181–1204
Peterson CL, Herskowitz I (1992) Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68:573–583
Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD (2005) Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell 123:233–248
Raisner RM, Madhani HD (2006) Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev 16:119–124
Ramachandran S, Zentner GE, Henikoff S (2015) Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res 25:381–390
Rando OJ, Winston F (2012) Chromatin and transcription in yeast. Genetics 190:351–387
Rawal Y, Chereji RV, Qiu HF, Ananthakrishnan S, Govind CK, Clark DJ, Hinnebusch AG (2018) SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast. Gene Dev 32:695–710
Rhee HS, Bataille AR, Zhang L, Pugh BF (2014) Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 159:1377–1388
Roberts CW, Orkin SH (2004) The SWI/SNF complex–chromatin and cancer. Nat Rev Cancer 4:133–142
Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T (2011) The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J 30:2596–2609
Shen X, Mizuguchi G, Hamiche A, Wu C (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541–544
Shetty A, Lopes JM (2010) Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryot Cell 9:1845–1855
Shimizu M, Takahashi K, Lamb TM, Shindo H, Mitchell AP (2003) Yeast Ume6p repressor permits activator binding but restricts TBP binding at the HOP1 promoter. Nucleic Acids Res 31:3033–3037
Singleton MR, Dillingham MS, Wigley DB (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76:23–50
Smolle MM (2018) Chd1 bends over backward to remodel. Nat Struct Mol Biol 25:2–3
Strathern JN, Klar AJ, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C (1982) Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31:183–192
Sudarsanam P, Cao Y, Wu L, Laurent BC, Winston F (1999) The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J 18:3101–3106
Sugiyama M, Nikawa J (2001) The Saccharomyces cerevisiae Isw2p-Itc1p complex represses INO1 expression and maintains cell morphology. J Bacteriol 183:4985–4993
Sundaramoorthy R (2019) Nucleosome remodelling: structural insights into ATP-dependent remodelling enzymes. Essays Biochem 63:45–58
Sundaramoorthy R, Hughes AL, El-Mkami H, Norman DG, Ferreira H, Owen-Hughes T (2018) Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. eLife 7:35720
Syntichaki P, Topalidou I, Thireos G (2000) The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404:414–417
Tirosh I, Barkai N (2008) Two strategies for gene regulation by promoter nucleosomes. Genome Res 18:1084–1091
Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner KP (2013) Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154:1207–1219
Tsukiyama T, Daniel C, Tamkun J, Wu C (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83:1021–1026
Udugama M, Sabri A, Bartholomew B (2011) The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol 31:662–673
Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598–602
Vary JC, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T (2003) Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol 23:80–91
Voutsina A, Fragiadakis GS, Gkouskou K, Alexandraki D (2019) Synergy of Hir1, Ssn6, and Snf2 global regulators is the functional determinant of a Mac1 transcriptional switch in S. cerevisiae copper homeostasis. Curr Genet 65:799–816
Watanabe S, Tan D, Lakshminarasimhan M, Washburn MP, Hong EJ, Walz T, Peterson CL (2015) Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat Commun 6:7108
Watson JD, Gann A, Bt A, Levine M, Bs P, Losick R (2014) Molecular biology of the gene, 7th edn. CSHL Press, Cold Spring Harbor
Whitehouse I, Rando OJ, Delrow J, Tsukiyama T (2007) Chromatin remodelling at promoters suppresses antisense transcription. Nature 450:1031–U1033
Xella B, Goding C, Agricola E, Di Mauro E, Caserta M (2006) The ISWI and CHD1 chromatin remodelling activities influence ADH2 expression and chromatin organization. Mol Microbiol 59:1531–1541
Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ (2011) Structure and mechanism of the chromatin remodelling factor ISW1a. Nature 472:448–453
Yan L, Chen Z (2020) A unifying mechanism of DNA translocation underlying chromatin remodeling. Trends Biochem Sci 45:217–227
Ye Y, Wu H, Chen K, Clapier CR, Verma N, Zhang W, Deng H, Cairns BR, Gao N, Chen Z (2019) Structure of the RSC complex bound to the nucleosome. Science 366:838–843
Zhang F, Kirouac M, Zhu N, Hinnebusch AG, Rolfes RJ (1997) Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol Cell Biol 17:3272–3283
Zhang H, Roberts DN, Cairns BR (2005) Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219–231
Zhang Z, Wang X, Xin J, Ding Z, Liu S, Fang Q, Yang N, Xu RM, Cai G (2018) Architecture of SWI/SNF chromatin remodeling complex. Protein Cell 9:1045–1049
Zhou CY, Johnson SL, Lee LJ, Longhurst AD, Beckwith SL, Johnson MJ, Morrison AJ, Narlikar GJ (2018) The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol Cell 69:677–688
Zimmer C, Fabre E (2019) Chromatin mobility upon DNA damage: state of the art and remaining questions. Curr Genet 65:1–9
Acknowledgements
The authors wish to thank laboratory members for discussion and Michelle Hanna for proofreading. This work was supported by the Natural Sciences and Engineering Research Council of Canada Discovery Grant RGPIN-2014-04580 to WX.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lin, A., Du, Y. & Xiao, W. Yeast chromatin remodeling complexes and their roles in transcription. Curr Genet 66, 657–670 (2020). https://doi.org/10.1007/s00294-020-01072-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-020-01072-0