Abstract
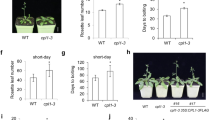
The plant circadian clock regulates a number of central activities such as growth, flowering, abiotic and biotic stress responses, and metabolism. The PSEUDO-RESPONSE REGULATORs (PRRs) contribute to the regulation of the circadian oscillator and clock output processes. In order to elucidate the role of Oncidium PRR9, we isolated and characterized a PRR9 homolog (OnPRR9) from commercially important Oncidium ‘Gower Ramsey’. OnPRR9 encodes a 603-amino acid protein with a N-terminal REC domain and a C-terminal CCT domain. Phylogenic analysis displayed that the OnPRR9 was orthologous to other known or deduced PRR proteins at the sequence level. Examination of the transcriptional expression profile of the OnPRR9 revealed that it is subjected to circadian rhythms under the conditions of 12-h light and 12-h dark cycles (12:12LD) and continuous light (LL) but not the continuous dark (DD) conditions. Genetic complementation showed that it can partially rescue the flowering time defect of Arabidopsis mutant prr5/9 grown under the long-day (16L:8D) conditions. Overexpression analysis indicated that the OnPRR9 gene can promote flowering and inhibit hypocotyl elongation under the short-day (8L:16D) conditions. Expression pattern of the OnPRR9 gene in petals during flower development and senescence implied that it may play a role in the initiation of flower senescence. These results extend our understanding of the functional conservation of PRR9 and may provide a means of regulating flowering time of Oncidium.







Similar content being viewed by others
References
Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11:36–42
Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293:880–883
Anwer MU, Boikoglou E, Herrero E, Hallstein M, Davis AM, Velikkakam James G, Nagy F, Davis SJ (2014) Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. ELife 3:e02206
Bendix C, Marshall CM, Harmon FG (2015) Circadian clock genes universally control key agricultural traits. Mol Plant 8:1135–1152
Bourret RB (2010) Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol 13:142–149
Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21:120–125
Endo M, Araki T, Nagatani A (2016) Tissue-specific regulation of flowering by photoreceptors. Cell Mol Life Sci 73:829–839
Farré EM, Liu T (2013) The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr Opin Plant Biol 16:621–629
Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15:47–54
Galperin MY (2006) Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188:4169–4182
Greenham K, McClung CR (2015) Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16:598–610
Harmer SL (2009) The circadian system in higher plants. Annu Rev Plant Biol 60:357–377
Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, Webb A, Goncalves J, Davis SJ (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24:428–443
Inoue K, Araki T, Endo M (2018) Circadian clock during plant development. J Plant Res 131:59–66
Johansson M, Köster T (2019) On the move through time—a historical review of plant clock research. Plant Biol (Stuttg) 21(Suppl 1):13–20
Kim JA, Yang TJ, Kim JS, Park JY, Kwon SJ, Lim MH, Jin M, Lee SC, Lee SI, Choi BS, Um SH, Kim HI, Chun C, Park BS (2007) Isolation of circadian-associated genes in Brassica rapa by comparative genomics with Arabidopsis thaliana. Mol Cells 23:145–153
Kim JA, Kim JS, Hong JK, Lee YH, Choi BS, Seol YJ, Jeon CH (2012) Comparative mapping, genomic structure, and expression analysis of eight pseudo-response regulator genes in Brassica rapa. Mol Genet Genomics 287:373–388
Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, Schoof H, Nagy F, Bujnicki JM, Davis SJ (2009) Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J 3:350–366
Kolmos E, Herrero E, Bujdoso N, Millar AJ, Tóth R, Gyula P, Nagy F, Davis SJ (2011) A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell 23:3230–3246
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: Insight into Histo-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41:791–803
Makino S, Matsushika A, Kojima M, Oda Y, Mizuno T (2001) Light response of the circadian waves of the APRR1/TOC1 quintet: when does the quintet start singing rhythmically in Arabidopsis? Plant Cell Physiol 42:334–339
Más P (2008) Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol 18:273–281
Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudoresponse regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41:1002–1012
Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T (2003) The evolutionarily conserved OsPRR quintet: rice pseudoresponse regulators implicated in circadian rhythm. Plant Cell Physiol 44:1229–1236
Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T (2005) Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci Biotechnol Biochem 69:410–414
Murakami M, Tago Y, Yamashino T, Mizuno T (2007) Characterization of the rice circadian clock-associated pseudo-response regulators in Arabidopsis thaliana. Biosci Biotechnol Biochem 71:1107–1110
Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T (2007) Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol 48:822–832
Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22:594–605
Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109:17123–17128
Nohales MA, Kay SA (2016) Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23:1061–1069
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyls growth. Nature 475:398–402
Ramírez-Carvajal GA, Morse AM, Davis JM (2008) Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol 177:77–89
Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDOREPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7:e1001350
Ronald J, Davis SJ (2017) Making the clock tick: the transcriptional landscape of the plant circadian clock. F1000Res 6:951
Rugnone ML, Faigon Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Mas P, Yanovsky MJ (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110:12120–12125
Sanchez SE, Kay SA (2016) The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harb Perspect Biol 8:a027748
Satbhai SB, Yamashino T, Mizuno T, Aoki S (2011a) Heterologous expression and functional characterization of a Physcomitrella Pseudo response regulator homolog, PpPRR2, in Arabidopsis. Biosci Biotechnol Biochem 75:786–789
Satbhai SB, Yamashino T, Okada R, Nomoto Y, Mizuno T, Tezuka Y, Itoh T, Tomita M, Otsuki S, Aoki S (2011b) Pseudo-response regulator (PRR) homologues of the moss Physcomitrella patens: insights into the evolution of the PRR family in land plants. DNA Res 18:39–52
Sawa M, Kay SA, Imaizumi T (2008) Photoperiodic flowering occurs under internal and external coincidence. Plant Signal Behav 3:269–271
Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93:1219–1229
Seo PJ, Más P (2014) Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26:79–87
Seo PJ, Más P (2015) STRESSing the role of the plant circadian clock. Trends Plant Sci 20:230–237
Shi L-S, Sun H, Tian X-Y, Wu F-H, Cai X-Q, Zhuang Y-F, Liu J-P (2016) Developmental stages of Oncidium Gower Ramsey flower. J Trop Bio 7(440–443):449
Shim JS, Kubota A, Imaizumi T (2017) Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173:5–15
Song YH, Ito S, Imaizumi T (2010) Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol 13:594–603
Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289:768–771
Takata N, Saito S, Saito CT, Uemura M (2010) Phylogenetic footprint of the plant clock system in angiosperms: evolutionary processes of pseudo-response regulators. BMC Evol Biol 10:126
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93:1207–1217
West AH, Stock AM (2001) Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26:369–376
Wu JF, Tsai HL, Joanito I, Wu YC, Chang CW, Li YH, Wang Y, Hong JC, Chu JW, Hsu CP, Wu SH (2016) LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat Commun 7:13181
Yang C-P, Hu J, Yan B-Y, Gong X-X, Tan Y-R, Gao X, Wang D, Zhang H, Liu J-P (2018) Cloning and expression analysis of circadian-associated gene PRR7 (Pseudo-Response Regulator 7) from Oncidium. Chin J Trop Crops 39:1570–1579
Yang C-P, Xia Z-Q, Hu J, Zhuang Y-F, Pan Y-W, Liu J-P (2019) Transcriptome analysis of Oncidium petals provides new insights into the initiation of petal senescence. J Hortic Sci Biotech 94:12–23
Zdepski A, Wang W, Priest H, Ali F, Alam M, Mockler T, Michael T (2008) Conserved daily transcriptional programs in Carica papaya. Trop Plant Biol 1:236–245
Zhang Y, Wang Y, Wei H, Li N, Tian W, Chong K, Wang L (2018) Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol Plant 11:326–337
Acknowledgements
This work was funded by the Basic and Applied Basic Research Plans (Natural Science Field) for High-level Talents of Hainan Province in 2019 (2019RC147), the National Natural Science Foundation of China (No. 31560573), the Postgraduate Research and Innovation Projects in Hainan Province in 2019 (Hby2019-14, Hys2019140), and the Postgraduate Research and Innovation Projects of Crop Discipline in Hainan University (ZWCX2018030).
Author information
Authors and Affiliations
Contributions
CPY and YRT performed the experiment and wrote the manuscript. JPL designed the whole experiment. BYY, XXG, DW, XG, HZ, PW, SJL, YW and LYZ contributed in transgenic experiment. YWP helped collecting the plant samples.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2020_611_MOESM2_ESM.jpg
Suppementary file Fig. S2. The conserved domains of the OnPRR9 obtained by the NCBI's conserved domain database (CDD) analysis. (jpg 78 kb)
10725_2020_611_MOESM3_ESM.jpg
Suppementary file Fig. S3. The OnPRR9 complementation lines were confirmed by PCR analysis at DNA level (A) and RNA level (B). A. The expected fragments of OnPRR9 + GFP were detected in all four putative transgenic Arabidopsis lines (1-4). No amplified PCR product was detected in non-transgenic prr5/9 mutant (5) and blank control (6). B. The expected transcripts of Hygromycin were detected in all three putative transgenic Arabidopsis lines (1-3). No amplified PCR product was detected in non-transgenic prr5/9 mutant (4) and blank control (5). (jpg 36 kb)
10725_2020_611_MOESM4_ESM.jpg
Suppementary file Fig. S4. The OnPRR9 overexpression lines were confirmed by PCR analysis at DNA level (A) and RNA level (B). A. The expected fragments of OnPRR9 + GFP were detected in all three putative transgenic Arabidopsis lines (1-3). No amplified PCR product was detected in non-transgenic wild type Arabidopsis (4) and blank control (5). B. The expected transcripts of the OnPRR9 were detected in all three putative transgenic Arabidopsis lines (1-3). No amplified PCR product was detected in non-transgenic wild type Arabidopsis (4). (jpg 34 kb)
Rights and permissions
About this article
Cite this article
Yang, CP., Tan, YR., Yan, BY. et al. Molecular characterization of clock-associated PSEUDO-RESPONSE REGULATOR 9 gene from Oncidium ‘Gower Ramsey’. Plant Growth Regul 91, 371–381 (2020). https://doi.org/10.1007/s10725-020-00611-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00611-6