Abstract
Background
Sal-like protein 4 (SALL4), an embryonic stem cell factor, has been reported to play an essential role in embryogenesis and oncogenesis. As yet, however, the expression and role of this transcription factor in head and neck squamous cell carcinoma (HNSCC) has not been established.
Methods
We assessed SALL4 mRNA expression in a well-characterised dataset of 230 HNSCC samples (test cohort 110 cases and validation cohort 120 cases). We also transfected HNSCC cells (FaDu and UM-SCC-6) with SALL4 siRNA and assessed its effects on proliferation and expression of specific epigenetic factors in order to uncover the role of SALL4 in HNSCC.
Results
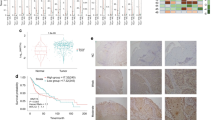
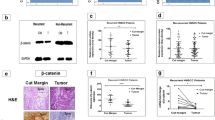
Overexpression of SALL4 was detected in tumour samples of both cohorts. HNSCC cells treated with SALL4 siRNA showed a reduction in growth and a decrease in DNA methyltransferase 3 alpha (DNMT3A) expression. In the patient cohorts, SALL4 overexpression was found to significantly correlate with disease recurrence (p < 0.001) and SALL4 methylation status (p = 0.002). We also found that DNMT3A was significantly upregulated upon SALL4 upregulation (p < 0.001). High expression levels of SALL4 correlated with decreases in disease-free survival (DFS) rates (log-rank test, p < 0.001). Multivariate analysis revealed that SALL4 expression served as an independent prognostic factor for DFS (hazard ratio: 2.566, 95% confidence interval: 1.598–4.121; p < 0.001).
Conclusions
Our findings indicate that SALL4 upregulation correlates with HNSCC tumour aggressiveness and an adverse patient outcome. Our findings also indicate that DNMT3A may synergistically contribute to the regulatory effects of SALL4. Our findings provide insight into SALL4-mediated HNSCC development via epigenetic modulation.




Similar content being viewed by others
Abbreviations
- SALL4:
-
Sal-like protein 4
- HNSCC:
-
head and neck squamous cell carcinoma
- DNMT3A:
-
DNA methyltransferase 3 alpha
- DFS:
-
disease-free survival
- ROC:
-
Receiver operating characteristic
- TCGA:
-
The Cancer Genome Atlas
- EGFR:
-
epidermal growth factor receptor
- qRT-PCR:
-
quantitative reverse transcription PCR
- Q-MSP:
-
quantitative methylation-specific PCR
References
M.R. Migden, D. Rischin, C.D. Schmults, A. Guminski, A. Hauschild, K.D. Lewis, C.H. Chung, L. Hernandez-Aya, A.M. Lim, A.L.S. Chang, G. Rabinowits, A.A. Thai, L.A. Dunn, B.G.M. Hughes, N.I. Khushalani, B. Modi, D. Schadendorf, B. Gao, F. Seebach, S. Li, J. Li, M. Mathias, J. Booth, K. Mohan, E. Stankevich, H.M. Babiker, I. Brana, M. Gil-Martin, J. Homsi, M.L. Johnson, V. Moreno, J. Niu, T.K. Owonikoko, K.P. Papadopoulos, G.D. Yancopoulos, I. Lowy, M.G. Fury, PD-1 Blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med 379, 341–351 (2018)
T. Kanazawa, K. Misawa, Y. Misawa, T. Uehara, H. Fukushima, G. Kusaka, M. Maruta, T.E. Carey, G-protein-coupled receptors: Next generation therapeutic targets in head and neck cancer? Toxins (Basel) 7, 2959–2984 (2015)
M. Mikulandra, A. Kobescak, B. Verillaud, P. Busson, T. Matijevic Glavan, Radio-sensitization of head and neck cancer cells by a combination of poly(I:C) and cisplatin through downregulation of survivin and c-IAP2. Cell Oncol 42, 29–40 (2019)
S. Roy, M. Kar, A. Saha, S. Padhi, B. Banerjee, Role of beta-catenin in cisplatin resistance, relapse and prognosis of head and neck squamous cell carcinoma. Cell Oncol 41, 185–200 (2018)
V. Budach, I. Tinhofer, Novel prognostic clinical factors and biomarkers for outcome prediction in head and neck cancer: A systematic review. Lancet Oncol 20, e313–e326 (2019)
J.Y. Hur, H.R. Kim, J.Y. Lee, S. Park, J.A. Hwang, W.S. Kim, S. Yoon, C.M. Choi, J.K. Rho, J.C. Lee, CDK7 inhibition as a promising therapeutic strategy for lung squamous cell carcinomas with a SOX2 amplification. Cell Oncol 42, 449–458 (2019)
P. Boscolo-Rizzo, M.C. Da Mosto, E. Rampazzo, S. Giunco, A. Del Mistro, A. Menegaldo, L. Baboci, M. Mantovani, G. Tirelli, A. De Rossi, Telomeres and telomerase in head and neck squamous cell carcinoma: From pathogenesis to clinical implications. Cancer Metastasis Rev 35, 457–474 (2016)
K. Misawa, Y. Misawa, T. Kanazawa, D. Mochizuki, A. Imai, S. Endo, T.E. Carey, H. Mineta, Epigenetic inactivation of galanin and GALR1/2 is associated with early recurrence in head and neck cancer. Clin Exp Metastasis 33, 187–195 (2016)
T. Nakagawa, K. Matsusaka, K. Misawa, S. Ota, K. Takane, M. Fukuyo, B. Rahmutulla, K.I. Shinohara, N. Kunii, D. Sakurai, T. Hanazawa, H. Matsubara, Y. Nakatani, Y. Okamoto, A. Kaneda, Frequent promoter hypermethylation associated with human papillomavirus infection in pharyngeal cancer. Cancer Lett 407, 21–31 (2017)
J. Kohlhase, R. Schuh, G. Dowe, R.P. Kuhnlein, H. Jackle, B. Schroeder, W. Schulz-Schaeffer, H.A. Kretzschmar, A. Kohler, U. Muller, M. Raab-Vetter, E. Burkhardt, W. Engel, R. Stick, Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene Spalt. Genomics 38, 291–298 (1996)
L. Yang, L. Liu, H. Gao, J.P. Pinnamaneni, D. Sanagasetti, V.P. Singh, K. Wang, M. Mathison, Q. Zhang, F. Chen, Q. Mo, T. Rosengart, J. Yang, The stem cell factor SALL4 is an essential transcriptional regulator in mixed lineage leukemia-rearranged leukemogenesis. J Hematol Oncol 10, 159 (2017)
H. Tatetsu, N.R. Kong, G. Chong, G. Amabile, D.G. Tenen, L. Chai, SALL4, the missing link between stem cells, development and cancer. Gene 584, 111–119 (2016)
K. Mei, A. Liu, R.W. Allan, P. Wang, Z. Lane, T.W. Abel, L. Wei, H. Cheng, S. Guo, Y. Peng, D. Rakheja, M. Wang, J. Ma, M.M. Rodriguez, J. Li, D. Cao, Diagnostic utility of SALL4 in primary germ cell tumors of the central nervous system: A study of 77 cases. Mod Pathol 22, 1628–1636 (2009)
K.J. Yong, C. Gao, J.S. Lim, B. Yan, H. Yang, T. Dimitrov, A. Kawasaki, C.W. Ong, K.F. Wong, S. Lee, S. Ravikumar, S. Srivastava, X. Tian, R.T. Poon, S.T. Fan, J.M. Luk, Y.Y. Dan, M. Salto-Tellez, L. Chai, D.G. Tenen, Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med 368, 2266–2276 (2013)
J. Yang, T.R. Corsello, Y. Ma, Stem cell gene SALL4 suppresses transcription through recruitment of DNA methyltransferases. J Biol Chem 287, 1996–2005 (2012)
J. Xiong, Z. Zhang, J. Chen, H. Huang, Y. Xu, X. Ding, Y. Zheng, R. Nishinakamura, G.L. Xu, H. Wang, S. Chen, S. Gao, B. Zhu, Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-Methylcytosine. Mol Cell 64, 913–925 (2016)
K. Misawa, D. Mochizuki, A. Imai, Y. Misawa, S. Endo, M. Mima, H. Kawasaki, T.E. Carey, T. Kanazawa, Epigenetic silencing of SALL3 is an independent predictor of poor survival in head and neck cancer. Clin Epigenetics 9, 64 (2017)
K. Misawa, Y. Misawa, A. Imai, D. Mochizuki, S. Endo, M. Mima, R. Ishikawa, H. Kawasaki, T. Yamatodani, T. Kanazawa, Epigenetic modification of SALL1 as a novel biomarker for the prognosis of early stage head and neck cancer. J Cancer 9, 941–949 (2018)
A. Imai, D. Mochizuki, Y. Misawa, T. Nakagawa, S. Endo, M. Mima, S. Yamada, H. Kawasaki, T. Kanazawa, K. Misawa, SALL2 is a novel prognostic methylation marker in patients with oral squamous carcinomas: Associations with SALL1 and SALL3 methylation status. DNA Cell Biol 38, 678–687 (2019)
D. Mochizuki, Y. Misawa, H. Kawasaki, A. Imai, S. Endo, M. Mima, S. Yamada, T. Nakagawa, T. Kanazawa, K. Misawa, Aberrant epigenetic regulation in head and neck Cancer due to distinct EZH2 overexpression and DNA Hypermethylation. Int J Mol Sci 19 (2018)
K. Misawa, D. Mochizuki, A. Imai, S. Endo, M. Mima, Y. Misawa, T. Kanazawa, T.E. Carey, H. Mineta, Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget 7, 26087–26098 (2016)
W.Y. Huang, S.D. Hsu, H.Y. Huang, Y.M. Sun, C.H. Chou, S.L. Weng, H.D. Huang, MethHC: A database of DNA methylation and gene expression in human cancer. Nucleic Acids Res 43, D856–D861 (2015)
J. Itou, W. Li, S. Ito, S. Tanaka, Y. Matsumoto, F. Sato, M. Toi, Sal-like 4 protein levels in breast cancer cells are post-translationally down-regulated by tripartite motif-containing 21. J Biol Chem 293, 6556–6564 (2018)
D. Cao, P.A. Humphrey, R.W. Allan, SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors. Cancer 115, 2640–2651 (2009)
C. Sun, P. Lan, Q. Han, M. Huang, Z. Zhang, G. Xu, J. Song, J. Wang, H. Wei, J. Zhang, R. Sun, C. Zhang, Z. Tian, Oncofetal gene SALL4 reactivation by hepatitis B virus counteracts miR-200c in PD-L1-induced T cell exhaustion. Nat Commun 9, 1241 (2018)
W. Du, L. Ni, B. Liu, Y. Wei, Y. Lv, S. Qiang, J. Dong, X. Liu, Upregulation of SALL4 by EGFR activation regulates the stemness of CD44-positive lung cancer. Oncogenesis 7, 36 (2018)
J.U. Marquardt, S.S. Thorgeirsson, Sall4 in "stemness"-driven hepatocarcinogenesis. N Engl J Med 368, 2316–2318 (2013)
S.S. Zeng, T. Yamashita, M. Kondo, K. Nio, T. Hayashi, Y. Hara, Y. Nomura, M. Yoshida, T. Hayashi, N. Oishi, H. Ikeda, M. Honda, S. Kaneko, The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol 60, 127–134 (2014)
L. Vilorio-Marques, V. Martin, C. Diez-Tascon, M.F. Gonzalez-Sevilla, T. Fernandez-Villa, E. Honrado, V. Davila-Batista, A.J. Molina, The role of EZH2 in overall survival of colorectal cancer: A meta-analysis. Sci Rep 7, 13806 (2017)
J. Itou, Y. Matsumoto, K. Yoshikawa, M. Toi, Sal-like 4 (SALL4) suppresses CDH1 expression and maintains cell dispersion in basal-like breast cancer. FEBS Lett 587, 3115–3121 (2013)
J. He, M. Zhou, X. Chen, D. Yue, L. Yang, G. Qin, Z. Zhang, Q. Gao, D. Wang, C. Zhang, L. Huang, L. Wang, B. Zhang, J. Yu, Y. Zhang, Inhibition of SALL4 reduces tumorigenicity involving epithelial-mesenchymal transition via Wnt/beta-catenin pathway in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 35, 98 (2016)
K. Nimura, K. Ura, H. Shiratori, M. Ikawa, M. Okabe, R.J. Schwartz, Y. Kaneda, A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to wolf-Hirschhorn syndrome. Nature 460, 287–291 (2009)
J. Yang, SALL4 as a transcriptional and epigenetic regulator in normal and leukemic hematopoiesis. Biomark Res 6, 1 (2018)
T. Baubec, D.F. Colombo, C. Wirbelauer, J. Schmidt, L. Burger, A.R. Krebs, A. Akalin, D. Schubeler, Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015)
J. Lu, H.W. Jeong, N. Kong, Y. Yang, J. Carroll, H.R. Luo, L.E. Silberstein, Yupoma, L. Chai, Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One 4, e5577 (2009)
A. Li, Y. Jiao, K.J. Yong, F. Wang, C. Gao, B. Yan, S. Srivastava, G.S. Lim, P. Tang, H. Yang, D.G. Tenen, L. Chai, SALL4 is a new target in endometrial cancer. Oncogene 34, 63–72 (2015)
X. Zhang, X. Yuan, W. Zhu, H. Qian, W. Xu, SALL4: An emerging cancer biomarker and target. Cancer Lett 357, 55–62 (2015)
Y. Ma, W. Cui, J. Yang, J. Qu, C. Di, H.M. Amin, R. Lai, J. Ritz, D.S. Krause, L. Chai, SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood 108, 2726–2735 (2006)
X. Yue, L. Xiao, Y. Yang, W. Liu, K. Zhang, G. Shi, H. Zhou, J. Geng, X. Ning, J. Wu, Q. Zhang, High cytoplasmic expression of SALL4 predicts a malignant phenotype and poor prognosis of breast invasive ductal carcinoma. Neoplasma 62, 980–988 (2015)
L. Zhang, Y. Yan, Y. Jiang, Y. Cui, Y. Zou, J. Qian, C. Luo, Y. Lu, X. Wu, The expression of SALL4 in patients with gliomas: High level of SALL4 expression is correlated with poor outcome. J Neuro-Oncol 121, 261–268 (2015)
S. Onder, O.C. Taskin, F. Sen, S. Topuz, S. Kucucuk, H. Sozen, R. Ilhan, S. Tuzlali, E. Yavuz, High expression of SALL4 and fascin, and loss of E-cadherin expression in undifferentiated/dedifferentiated carcinomas of the endometrium: An immunohistochemical and clinicopathologic study. Medicine (Baltimore) 96, e6248 (2017)
N. Yanagihara, D. Kobayashi, K. Kuribayashi, M. Tanaka, T. Hasegawa, N. Watanabe, Significance of SALL4 as a drugresistant factor in lung cancer. Int J Oncol 46, 1527–1534 (2015)
M. Miettinen, Z. Wang, P.A. McCue, M. Sarlomo-Rikala, J. Rys, W. Biernat, J. Lasota, Y.S. Lee, SALL4 expression in germ cell and non-germ cell tumors: A systematic immunohistochemical study of 3215 cases. Am J Surg Pathol 38, 410–420 (2014)
S. Zhou, R. Venkatramani, E. Gomulia, N. Shillingford, L. Wang, The diagnostic and prognostic value of SALL4 in hepatoblastoma. Histopathology 69, 822–830 (2016)
X. Zhang, P. Zhang, M. Shao, X. Zang, J. Zhang, F. Mao, H. Qian, W. Xu, SALL4 activates TGF-beta/SMAD signaling pathway to induce EMT and promote gastric cancer metastasis. Cancer Manag Res 10, 4459–4470 (2018)
Acknowledgments
The authors would like to thank Ms. Yuko Mohri for her excellent technical support and Editage (www.editage.jp) for English language editing.
Funding
This study was funded by a Grant-in-Aid for Scientific Research (No. 17 K11380, No. 17 K16903, 17 K16904, 19 K09866, 19 K09906 and 19 K18728) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Contributions
KM and YM conceived the study. KM, YM and KH designed the experiments. MM, YS, SI, DM, TK, TK, SE and MM analysed the data and prepared the figures and tables. All authors participated in writing the manuscript, reviewed its drafts, approved its final version and agreed with its submission.
Corresponding author
Ethics declarations
Conflict of interest
None to declare.
Ethics approval and consent to participate
The research methodology employed in this study was approved by The Institutional Review Board of the Hamamatsu University School of Medicine. All study subjects provided written informed consent.
Consent for publication
Consent for publication was obtained from all patients.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Fig. S1
Kaplan–Meier survival curves for patients with (A) oral cancer (n = 76; p = 0.026), (B) hypopharyngeal cancer (n = 54; p = 0.420), (C) laryngeal cancer (n = 51; p = 0.173), and (D) oropharyngeal cancer (n = 35; p = 0.111). * p < 0.05. (PNG 89 kb)
Supplementary Fig. S2
Data regarding SALL4 (A), DNMT3A (B), and DNMT3B (C) mRNA expression in head and neck squamous cell carcinoma were obtained from TCGA (https://tcga-data.nci.nih.gov/tcga/) and MethHC (http://methhc.mbc.nctu.edu.tw/php/index.php). * p < 0.05, ** p < 0.01. (PNG 94 kb)
Supplementary Fig. S3
Data regarding SALL4, CDH1 (A), COL1A2 (B) and CDH13 (C) mRNA expression in head and neck squamous cell carcinoma were obtained from TCGA (https://tcga-data.nci.nih.gov/tcga/) and MethHC (http://methhc.mbc.nctu.edu.tw/php/index.php). * p < 0.05. (PNG 109 kb)
Supplementary Table S1
(DOCX 20 kb)
Supplementary Table S2
(DOCX 19 kb)
Supplementary Table S3
(DOCX 20 kb)
Supplementary Table S4
(DOCX 18 kb)
Supplementary Table S5
(DOCX 21 kb)
Rights and permissions
About this article
Cite this article
Misawa, K., Misawa, Y., Mima, M. et al. Overexpression of Sal-like protein 4 in head and neck cancer: epigenetic effects and clinical correlations. Cell Oncol. 43, 631–641 (2020). https://doi.org/10.1007/s13402-020-00509-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-020-00509-5