Abstract
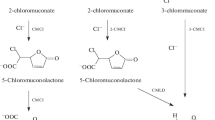
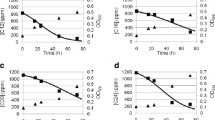
Naphthalene, as a component of crude oil, is a common environmental pollutant. Biochemical and genetic aspects of naphthalene catabolism have been examined in most detail in the bacteria of Pseudomonas genus. In pseudomonads, the key intermediate in naphthalene degradation is salicylate. In this study, we investigated the ability of Rhodococcus opacus strain 3D to utilize naphthalene as a sole carbon and energy source. The characteristic feature of this strain is the inability to grow in the mineral medium supplemented with salicylate (typical intermediate of naphthalene degradation in Gram-negative bacteria). The absence of salicylate hydroxylase activity and salicylate accumulation in the course of R. opacus 3D cultivation in the mineral medium supplemented with naphthalene indicated existence of an alternative pathway of naphthalene oxidation. At the same time, R. opacus 3D was able to use monoaromatic compounds (salts of gentisic, ortho-phthalic, and 2-hydroxycinnamic acids and coumarin) as growth substrates. Based on the analysis of enzymatic activities, identification of the reaction intermediates, genetic determinants, and growth substrates, we concluded that R. opacus 3D carries out naphthalene degradation through an alternative pathway via formation of ortho-phthalic acid, which is untypical for pseudomonads. Using mass spectrometry, we showed for the first time that salicylic acid associate formed in trace amounts in the process of naphthalene degradation is not further metabolized and accumulated in the growth medium in a form of a dimer.
Similar content being viewed by others
Abbreviations
- Cat 1,2-DO:
-
catechol 1,2- dioxygenase
- Cat 2,3-DO:
-
catechol 2,3-dioxygenase
- CFU:
-
colony-forming unit
- GDO:
-
gentisate dioxygenase
- MCI:
-
muconate cycloisomerase
- NDO:
-
naphthalene 1,2-dioxygenase
- PAH:
-
polycyclic aromatic hydrocarbon
- PCA:
-
protocatechuic acid
- PCA 3,4-DO:
-
pro-tocatechuate 3,4-dioxygenase
- S1H:
-
salicylate 1-hydroxylase
- TLC:
-
thin-layer chromatography
References
Juhasz, A. L., and Naidu, R. (2000) Bioremediation of high molecular weight PAHs: a review of the microbial degradation of benzo[a]pyrene, Int. Biodeterior. Biodegrad., 45, 57–88; doi: https://doi.org/10.1016/S0964-8305(00)00052-4.
Fonger, G. C., Hakkinen, P., Jordan, S., and Publicker, S. (2014) The National Library of Medicine’s (NLM) Hazardous Substances Data Bank (HSDB): background, recent enhancements and future plans, Toxicology, 325, 209–216; doi: https://doi.org/10.1016/j.tox.2014.09.003.
Davies, J. I., and Evans, W. C. (1964) Oxidative metabolism of naphthalene nucleus by soil pseudomonads — ring fission mechanism, Biochem. J., 91, 251–261; doi: https://doi.org/10.1042/bj0910251.
Jeffrey, A. M., Yeh, H. J., Jerina, D. M., Patel, T. R., Davey, J. F., and Gibson, D. T. (1975) Initial reactions in the oxidation of naphthalene by Pseudomonas putida, Biochemistry, 14, 575–584; doi: https://doi.org/10.1021/bi00674a018
Patel, T. R., and Barnsley, E. A. (1980) Naphthalene metabolism by pseudomonads: purification and properties of 1,2-dihydroxynaphthalene dioxygenase, J. Bacteriol., 143, 668–673.
Assinder, S. J., and Williams, P. A. (1988) Comparison of the meta pathway operons on NAH plasmid pWW60-22 and TOL plasmid pWW53-4 and its evolutionary significance, J. Gen. Microbiol., 134, 2769–2778; doi: https://doi.org/10.1099/00221287-134-10-2769.
Rossello-Mora, R. A., Lalucat, J., and Garcia-Valdes, E. (1994) Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains, Appl. Environ. Microbiol., 60, 966–972.
Tay, M., Roizman, D., Cohen, Y., Tolker-Nielsen, T., Givskov, M., and Yang, L. (2014) Draft genome sequence of the model naphthalene-utilizing organism Pseudomonas putida OUS82, Genome Announc., 2, e01161–13; doi: https://doi.org/10.1128/genomeA.01161-13
Yen, K. M., and Serdar, C. M. (1988) Genetics of naphthalene catabolism in pseudomonads, Crit. Rev. Microbiol., 15, 247–268.
Kochetkov, V. V., Balakshina, V. V., Mordukhova, E. A., and Boronin, A. M. (1997) Plasmids encoding naphthalene biodegradation in rhizosphere bacteria of the Pseudomonas genus, Mikrobiologiya, 66, 211–216.
Sevastsyanovich, Y. R., Krasowiak, R., Bingle, L. E. H., Haines, A. S., Sokolov, S. L., Kosheleva, I. A., Leuchuk, A. A., Titok, M. A., Smalla, K., and Thomas, C. M. (2008) Diversity of IncP-9 plasmids of Pseudomonas, Microbiology, 154, 2929–2941; doi: https://doi.org/10.1099/mic.0.2008/017939-0
Kiyohara, H., Torigoe, S., Kaida, N., Asaki, T., Iida, T., Hayashi, H., and Takizawa, N. (1994) Cloning and characterization of a chromosomal gene cluster, pah, that encodes the upper pathway for phenanthrene and naphthalene utilization by Pseudomonas putida OUS82, J. Bacteriol., 176, 2439–2443; doi: https://doi.org/10.1128/jb.176.8.2439-2443.1994.
Meyer, S., Moser, R., Neef, A., Stahl, U., and Kampfer, P. (1999) Differential detection of key enzymes of polyaromatic-hydrocarbon-degrading bacteria using PCR and gene probes, Microbiology, 145, 1731–1741; doi: https://doi.org/10.1099/13500872-145-7-1731.
Moser, R., and Stahl, U. (2001) Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria, Appl. Microbiol. Biotechnol., 55, 609–618; doi: https://doi.org/10.1007/s002530000489.
Kauppi, B., Lee, K., Carredano, E., Parales, R. E., and Gibson, D. T. (1998) Structure of an aromatic ring-hydroxylating dioxygenase naphthalene 1,2-dioxygenase, Structure, 6, 571–586; doi: https://doi.org/10.1016/s0969-2126(98)00059-8.
Parales, R. E., Parales, J. V., and Gibson, D. T. (1999) Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity, J. Bacteriol., 181, 1831–1837.
Bosch, R., Moore, E. R., Garcia-Valdes, E., and Pieper, D. H. (1999) NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10, J. Bacteriol., 181, 2315–2322.
Eaton, R. W., and Chapman, P. J. (1992) Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions, J. Bacteriol., 174, 7542–7554; doi: https://doi.org/10.1128/jb.174.23.7542-7554.1992.
Fuenmayor, S. L., Wild, M., Boyes, A. L., and Williams, P. A. (1998) A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2, J. Bacteriol., 180, 2522–2530.
Hickey, W. J., Sabat, G., Yuroff, A. S., Arment, A. R., and Perez-Lesher, J. (2001) Cloning, nucleotide sequencing, and functional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa strain JB2, Appl. Environ. Microbiol., 67, 4603–4609; doi: https://doi.org/10.1128/aem.67.10.4603-4609.2001.
Lee, J., Min, K. R., Kim, Y. C., Kim, C. K., Lim, J. Y., Yoon, H., Min, K. H., Lee, K. S., and Kim, Y. (1995) Cloning of salicylate hydroxylase gene and catechol 2,3-dioxygenase gene and sequencing of an intergenic sequence between the two genes of Pseudomonas putida KF715, Biochem. Biophys. Res. Commun., 211, 382–388; doi: https://doi.org/10.1006/bbrc.1995.1825.
Feng, Y., Khoo, H. E., and Poh, C. L. (1999) Purification and characterization of gentisate 1,2-dioxygenases from Pseudomonas alcaligenes NCIB 9867 and Pseudomonas putida NCIB 9869, Appl. Environ. Microbiol., 65, 946–950.
Chua, C. H., Feng, Y., Yeo, C. C., Khoo, H. E., and Poh, C. L. (2001) Identification of amino acid residues essential for catalytic activity of gentisate 1,2-dioxygenase from Pseudomonas alcaligenes NCIB 9867, FEMS Microbiol. Lett., 204, 141–146; doi: https://doi.org/10.1007/s10529-007-9421-7.
Zhou, N. Y., Fuenmayor, S. L., and Williams, P. A. (2001) nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism, J. Bacteriol., 183, 700–708; doi: https://doi.org/10.1128/JB.183.2.700-708.2001.
Goetz, F. E., and Harmuth, L. J. (1992) Gentisate pathway in Salmonella typhimurium: metabolism of m-hydroxybenzoate and gentisate, FEMS Microbiol. Lett., 76, 45–49; doi: https://doi.org/10.1016/0378-1097(92)90361-q.
Civilini, M., de Bertoldi, M., and Tell, G. (1999) Molecular characterization of Pseudomonas aeruginosa 2NR degrading naphthalene, Lett. Appl. Microbiol., 29, 181–186; doi: https://doi.org/10.1046/j.1365-2672.1999.00613.x.
Starovoitov, I. I., Nefedova, M. Yu., Yakovlev, G. I., Zyakun, A. M., and Adanin, V. M. (1975) Gentisic acid — product of microbial naphthalene oxidation, Izv. Akad. Nauk, Ser. Khim., 9, 2091–2092.
Monticello, D. J., Bakker, D., Schell, M., and Finnerty, W. R. (1985) Plasmid-borne TnS insertion mutations resulting in accumulation of gentisate from salicylate, Appl. Environ. Microbiol., 49, 761–764.
Fu, W., and Oriel, P. (1998) Gentisate 1,2-dioxygenase from Haloferax sp. D1227, Extremophiles, 2, 439–446; doi: https://doi.org/10.1007/s007920050090.
Werwath, J., Arfmann, H. A., Pieper, D. H., Timmis, K. N., and Wittich, R. M. (1998) Biochemical and genetic characterization of a gentisate 1,2-dioxygenase from Sphingomonas sp. strain RW5, J. Bacteriol., 180, 4171–4176.
Uz, I., Duan, Y. P., and Ogram, A. (2000) Characterization of the naphthalene-degrading bacterium Rhodococcus opacus M213, FEMS Microbiol. Lett., 185, 231–238; doi: https://doi.org/10.1111/j.1574-6968.2000.tb09067.x.
Auffret, M., Labbe, D., Thouand, G., Greer, C. W., and Fayolle-Guichard, F. (2009) Degradation of a mixture of hydrocarbons, gasoline, and diesel oil additives by Rhodococcus aetherivorans and Rhodococcus wratislaviensis, Appl. Environ. Microbiol., 75, 7774–7782; doi: https://doi.org/10.1128/AEM.01117-09
Larkin, M. J., Kulakov, L. A., and Allen, C. C. (2005) Biodegradation and Rhodococcus — masters of catabolic versatility, Curr. Opin. Biotechnol., 16, 282–290; doi: https://doi.org/10.1016/j.copbio.2005.04.007.
Dong, L., Nakashima, N., Tamura, N., and Tamura, T.. (2004) Isolation and characterization of the Rhodococcus opacus thiostrepton-inducible genes tipAL and tipAS: application for recombinant protein expression in Rhodococcus, FEMS Microbiol. Lett., 237, 35–40; doi: https://doi.org/10.1016/j.femsle.2004.06.012
Kulakov, L. A., Chen, S., Allen, C. C., and Larkin, M. J. (2005) Web-type evolution of Rhodococcus gene clusters associated with utilization of naphthalene, Appl. Environ. Microbiol., 71, 1754–1764; doi: https://doi.org/10.1128/AEM.71.4.1754-1764.2005.
Grund, E., Denecke, B., and Eichenlaub, R. (1992) Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4, Appl. Environ. Microbiol., 58, 1874–1877.
Allen, C., Boyd, D., and Larkin, M. (1997) Metabolism of naphthalene, 1-naphthol, indene, and indole by Rhodococcus sp. strain NCIMB 12038, Appl. Environ. Microbiol., 63, 151–155.
Larkin, M. J., and Day, M. J. (1986) The metabolism of carbaryl by three bacterial isolates, Pseudomonas spp. (NCBI12042 and 12043) and Rhodococcus sp. (NCIB12038) from garden soil, J. Appl. Bacteriol., 60, 233–242; doi: https://doi.org/10.1111/j.1365-2672.1986.tb01078.x.
Gorlatov, S. N., Mal’tseva, O. V., Shevchenko, V. L., and Golovleva, L. A. (1989) Degradation of chlorophenols by the Rhodococcus erythropolis culture, Mikrobiologiya, 58, 802–806.
Chernyavskaya, M. I. (2016) Characterization of strains of naphthalene-utilizing bacteria of the Rhodococcus genus, Works BSU. Physiol. Biochem. Mol. Basis Funct. Biosyst., 11, 190–197.
Plotnikova, E. G., Rybkina, D. O., Anan’ina, L. N., Yastrebova, O. V., and Demakov, V. A. (2006) Characteristics of microorganisms isolated from technogenic soil in Prikam’ye, Ekologiya, 4, 261–268.
Izmalkova, T. Yu., Gafarov, A. B., Sazonova, O. I., Sokolov, S. L., Kosheleva, I. A., and Boronin, A. M. (2018) Diversity of oil-degrading microorganisms in the Gulf of Finland (Baltic Sea) in spring and in summer, Microbiology, 87, 261–271; doi: https://doi.org/10.1134/S0026261718020054.
Anokhina, T. O., Volkova, O. V., Puntus, I. F., Filonov, A. E., Kochetkov, V. V., and Boronin, A. M. (2006) Plant growth-promoting Pseudomonas bearing catabolic plasmids: naphthalene degradation and effect on plants, Process Biochem., 41, 2417–2423; doi: https://doi.org/10.1016/j.procbio.2006.06.026.
Lane, D. J. (1991) 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematic (Stackebrandt, E., and Goodfellow, M., eds.) John Wiley and Sons, NY, pp. 115–175.
Subashchandrabose, S. R., Venkateswarlu, K., Naidu, R., and Megharaj, M. (2019) Biodegradation of high-molecular-weight PAHs by Rhodococcus wratislaviensis strain 9: overexpression of amidohydrolase induced by pyrene and BaP, Sci. Total Environ., 651, 813–821; doi: https://doi.org/10.1007/978-3-030-24035-63.
Choi, K. Y., Kim, D., Sul, W. J., Chae, J. C., Zylstra, G. J., Kim, Y. M., and Kim, E. (2005) Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcus sp. strain DK17, FEMS Microbiol. Lett., 252, 207–213; doi: https://doi.org/10.1016/j.femsle.2005.08.045.
Kulakov, L. A., Delcroix, V. A., Larkin, M. J., Ksenzenko, V. N., and Kulakova, A. N. (1998) Cloning of new Rhodococcus extradiol dioxygenase genes and study of their distribution in different Rhodococcus strains, Microbiology, 144, 955–963; doi: https://doi.org/10.1099/00221287-144-4-955.
Li, C., Zhang, C., Song, G., Liu, H., Sheng, G., Ding, Z., Wang, Z., Sun, Y., Xu, Y., and Chen, J. (2016) Characterization of a protocatechuate catabolic gene cluster in Rhodococcus ruber OA1 involved in naphthalene degradation, Ann. Microbiol., 66, 469–478; doi: https://doi.org/10.1007/s13213-015-1132-z.
Solyanikova, I. P., Borzova, O. V., Emel’yanova, E. V., Shumkova, E. S., Prisyazhnaya, N. V., Plotnikova, N. V., and Golovleva, L. A. (2016) Dioxygenases of chlorobiphenyl-degrading species Rhodococcus wratislaviensis G10 and chlorophenol-degrading species induced in benzoate-grown cells and genes potentially involved in these processes, Biochemistry (Moscow), 81, 986–998; doi: https://doi.org/10.1134/S000629791609008X.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Res., 22, 4673–4680; doi: https://doi.org/10.1093/nar/22.22.4673.
Dua, R. D., and Meera, S. (1981) Purification and characterization of naphthalene oxygenase from Corynebacterium renale, Eur. J. Biochem., 120, 461–465; doi: https://doi.org/10.1111/j.1432-1033.1981.tb05724.x
Shamsuzzaman, K. M., and Barnsley, E. A. (1974) The regulation of naphthalene oxygenase in pseudomonads, J. Gen. Microbiol., 83, 165–170; doi: https://doi.org/10.1099/00221287-83-1-165.
Hegeman, G. D. (1966) Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type, J. Bacteriol., 91, 1140–1154.
Crawford, R. L., Hutton, S. W., and Chapman, P. J. (1975) Purification and properties of gentisate 1,2-dioxygenase from Moraxella osloensis, J. Bacteriol., 121, 794–799.
Fujisawa, H., and Hayaishi, O. (1968) Protocatechuate 3,4-dioxygenase. I. Crystallization and characterization, J. Biol. Chem., 243, 2673–2681.
Schlomann, M., Schmidt, E., and Knackmuss, H. J. (1990) Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria, J. Bacteriol., 172, 5112–5118; doi: https://doi.org/10.1128/jb.172.9.5112-5118.1990.
Kulakova, A. N. (1988) Nature of Genetic Control of Naphthalene and Salicylic Acid Catabolism in the Pseudomonas putida BSA202 Strain: Candidate dissertation [in Russian], Institute of Biochemistry and Physiology of Microorganisms, Pushchino.
Pathak, A., Chauhan, A., Blom, J., Indest, K. J., Jung, C. M., Stothard, P., Bera, G., Green, S. J., and Ogram, A. (2016) Comparative genomics and metabolic analysis reveals peculiar characteristics of Rhodococcus opacus strain M213 particularly for naphthalene degradation, PLoS One, 11, e0161032; doi: https://doi.org/10.1371/journal.pone.0161032
Martinkova, L., Uhnakova, B., Patek, M., Nesvera, J., and Kren, V. (2009) Biodegradation potential of the genus Rhodococcus, Environ. Int., 35, 162–177; doi: https://doi.org/10.1016/j.envint.2008.07.018
McLeod, M. P., Warren, R. L., Hsiao, W. W., Araki, N., Myhre, M., Fernandes, C., Miyazawa, D., Wong, W., Lillquist, A. L., Wang, D., Dosanjh, M., Hara, H., Petrescu, A., Morin, R. D., Yang, G., Stott, J. M., Schein, J. E., Shin, H., Smailus, D., Siddiqui, A. S., Marra, M. A., Jones, S. J., Holt, R., Brinkman, F. S., Miyauchi, K., Fukuda, M., Davies, J. E., Mohn, W. W., and Eltis, L. D. (2006) The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse, Proc. Natl. Acad. Sci. USA, 103, 15582–15587; doi: https://doi.org/10.1073/pnas.0607048103.
Kulakova, A. N., Reid, K. A., Larkin, M. J., Allen, C. C., and Kulakov, L. A. (1996) Isolation of Rhodococcus rhodochrous NCIMB 13064 derivatives with new biodegradative abilities, FEMS Microbiol. Lett., 145, 227–231; doi: https://doi.org/10.1111/j.1574-6968.1996.tb08582.x.
Anastasi, E., MacArthur, I., Scortti, M., Alvarez, S., Giguere, S., and Vazquez-Boland, J. A. (2016) Pangenome and phylogenomic analysis of the pathogenic actinobacterium Rhodococcus equi, Genome Biol. Evol., 8, 3140–3148; doi: https://doi.org/10.1007/s00203-019-01695-z
Annweiler, E., Richnow, H. H., Antranikian, G., Hebenbrock, S., Garms, C., Franke, S., Francke, W., and Michaelis, W. (2000) Naphthalene degradation and incorporation of naphthalene-derived carbon into biomass by the thermophile Bacillus thermoleovorans, Appl. Environ. Microbiol., 66, 518–523; doi: https://doi.org/10.1128/aem.66.2.518-523.2000
Bubinas, A., Giedraityte, G., Kalediene, L., Nivinskiene, O., and Butkiene, R. (2008) Degradation of naphthalene by thermophilic bacteria via a pathway through protocatechuic acid, Cent. Eur. J. Biol., 3, 61–68; doi: https://doi.org/10.2478/s11535-007-0042-x.
Van Herwijnen, R., Springael, D., Slot, P., Govers, H. A., and Parsons, J. A. (2003) Degradation of anthracene by Mycobacterium sp. strain LB501T proceeds via a novel pathway, through o-phthalic acid, Appl. Environ. Microbiol., 69, 186–190; doi: https://doi.org/10.1128/aem.69.1.186-190.2003.
Acknowledgements
The authors express their gratitude to Dr. M. I. Chernyavskaya (Belarusian State University, Minsk, Republic of Belarus) for kindly providing R. pyridinivorans 5Ap (L5A-BSU) strain and to Dr. E. G. Plotnikova (Institute of Ecology and Genetics of Microorganisms, Ural Division of the Russian Academy of Sciences, Perm, Russia) for kindly providing R. wratislaviensis G10 strain.
Author information
Authors and Affiliations
Corresponding author
Additional information
Russian Text © The Author(s), 2020, published in Biokhimiya, 2020, Vol. 85, No. 3, pp. 412–427.
Funding
This work was supported by the Russian Foundation for Basic Research (project 18-34-00964).
Conflict of interest
The authors declare no conflict of interest in financial or any other sphere.
Ethical approval
This article does not contain description of studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Anokhina, T.O., Esikova, T.Z., Gafarov, A.B. et al. Alternative Naphthalene Metabolic Pathway Includes Formation of ortho-Phthalic Acid and Cinnamic Acid Derivatives in the Rhodococcus opacus Strain 3D. Biochemistry Moscow 85, 355–368 (2020). https://doi.org/10.1134/S0006297920030116
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920030116