Abstract
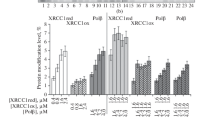
Base excision repair (BER) ensures correction of most abundant DNA lesions in mammals. The efficiency of this multistep DNA repair process that can occur via different pathways depends on the coordinated action of enzymes catalyzing its individual steps. The scaffold XRCC1 (X-ray repair cross-complementing protein 1) protein plays an important coordinating role in the repair of damaged bases and apurinic/apyrimidinic (AP) sites via short-patch (SP) BER pathway, as well as in the repair of single-strand DNA breaks. In this study, we demonstrated for the first time in vitro formation of the ternary XRCC1 complex with the key enzymes of SP BER — DNA polymerase β (Polβ) and DNA ligase IIIa (LiglIIa) — using the fluorescence-based technique. It was found that Polβ directly interacts with LiglIIa, but their complex is less stable than the XRCC1—Polβ and XRCC1—LigIIIa complexes. The effect of XRCC1 oxidation and composition of the multiprotein complex on the efficiency of DNA synthesis and DNA ligation during DNA repair has been explored. We found that formation of the disulfide bond between Cys12 and Cys20 residues as a result of XRCC1 oxidation (previously shown to modulate the protein affinity for Polβ), affects the yield of the final product of SP BER and of non-ligated DNA intermediates (substrates of long-patch BER). The effect of XRCC1 oxidation on the final product yield depended on the presence of AP endonuclease 1. Together with the data from our previous work, the results of this study suggest an important role of XRCC1 oxidation in the fine regulation of formation of BER complexes and their functional activity.
Similar content being viewed by others
Abbreviations
- APE1:
-
AP endonuclease 1
- AP site:
-
apurinic/apyrimidinic site
- dRp:
-
deoxyribose phosphate
- DTT:
-
dithiothreitol
- FAM:
-
5(6)-carboxyfluorescein
- FEN1:
-
flap endonuclease 1
- gap-DNA:
-
DNA duplex with one-nucleotide gap
- LigI/LigIIIa:
-
DNA ligase I/IIIa
- LP BER:
-
long-patch base excision repair
- nick-DNA:
-
DNA duplex with single-strand break
- PARP1:
-
poly(ADP-ribose) polymerase 1
- Polβ/PolS/Pole:
-
DNA polymerase β/S/e
- SP BER:
-
short-patch base excision repair
- XRCC1:
-
X-ray repair cross-complementing protein 1
- XRCC1ox:
-
oxidized form of XRCC1 protein
References
Abbotts, R., and Wilson, D. M., 3rd (2017) Coordination of DNA single strand break repair, Free Radic. Biol. Med., 107, 228–244, doi: https://doi.org/10.1016/j.freeradbiomed.2016.11.039.
Whitaker, A. M., Schaich, M. A., Smith, M. R., Flynn, T. S., and Freudenthal, B. D. (2017) Base excision repair of oxidative DNA damage: from mechanism to disease, Front. Biosci. (Landmark Ed.), 22, 1493–1522, doi: https://doi.org/10.2741/4555.
Caldecott, K. W. (2014) DNA single-strand break repair, Exp. Cell. Res., 329, 2–8, doi: https://doi.org/10.1016/j.yexcr.2014.08.027.
London, R. E. (2015) The structural basis of XRCC1-mediated DNA repair, DNA Repair (Amst.), 30, 90–103, doi: https://doi.org/10.1016/j.dnarep.2015.02.005.
Moor, N. A., and Lavrik, O. I. (2018) Protein—protein interactions in DNA base excision repair, Biochemistry (Moscow), 83, 411–422, doi: https://doi.org/10.1134/S0006297918040120.
Dutta, A., Yang, C., Sengupta, S., Mitra, S., and Hegde, M. L. (2015) New paradigms in the repair of oxidative damage in human genome: mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins, Cell. Mol. Life Sci., 72, 1679–1698, doi: https://doi.org/10.1007/s00018-014-1820-z.
Caldecott, K. W., McKeown, C. K., Tucker, J. D., Ljungquist, S., and Thompson, L. H. (1994) An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III, Mol. Cell. Biol., 14, 68–76, doi: https://doi.org/10.1128/mcb.14.1.68.
Kubota, Y., and Horiuchi, S. (2003) Independent roles of XRCC1’s two BRCT motifs in recovery from methylation damage, DNA Repair (Amst.), 2, 407–415, doi: https://doi.org/10.1016/s1568-7864(02)00242-2.
Parsons, J. L., Dianova, I. I., Allinson, S. L., and Dianov, G. L. (2005) DNA polymerase beta promotes recruitment of DNA ligase III alpha-XRCC1 to sites of base excision repair, Biochemistry, 44, 10613–10619, doi: https://doi.org/10.1021/bi050085m.
Moor, N. A., Vasil’eva, I. A., Anarbaev, R. O., Antson, A. A., and Lavrik, O. I. (2015) Quantitative characterization of protein—protein complexes involved in base excision DNA repair, Nucleic Acids Res., 43, 6009–6022, doi: https://doi.org/10.1093/nar/gkv569.
Kubota, Y., Nash, R. A., Klungland, A., Schär, P., Barnes, D. E., and Lindahl, T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein, EMBO J., 15, 6662–6670, doi: https://doi.org/10.1002/J.1460-2075.1996.tb01056.x
Wong, H. K., and Wilson, D. M., 3rd (2005) XRCC1 and DNA polymerase beta interaction contributes to cellular alkylating agent resistance and single-strand break repair, J. Cell. Biochem., 95, 794–804, doi: https://doi.org/10.1002/jcb.20448.
Fang, Q., Inanc, B., Schamus, S., Wang, X. H., Wei, L., Brown, A. R., Svilar, D., Sugrue, K. F., Goellner, E. M., Zeng, X., Yates, N. A., Lan, L., Vens, C., and Sobol, R. W. (2014) HSP90 regulates DNA repair via the interaction between XRCC1 and DNA polymerase β, Nat. Commun., 5, 5513, doi: https://doi.org/10.1038/ncomms6513.
Horton, J. K., Stefanick, D. F., Gassman, N. R., Williams, J. G., Gabel, S. A., Cuneo, M. J., Prasad, R., Kedar, P. S., Derose, E. F., Hou, E. W., London, R. E., and Wilson, S. H. (2013) Preventing oxidation of cellular XRCC1 affects PARP-mediated DNA damage responses, DNA Repair (Amst.), 12, 774–785, doi: https://doi.org/10.1016/j.dnarep.2013.06.004.
Horton, J. K., Seddon, H. J., Zhao, M. L., Gassman, N. R., Janoshazi, A. K., Stefanick, D. F., and Wilson, S. H. (2017) Role of the oxidized form of XRCC1 in protection against extreme oxidative stress, Free Radic. Biol. Med., 107, 292–300, doi: https://doi.org/10.1016/j.freeradbiomed.2017.02.005.
Kumar, A., Widen, S. G., Williams, K. R., Kedar, P., Karpel, R. L., and Wilson, S. H. (1990) Studies of the domain structure of mammalian DNA polymerase beta. Identification of a discrete template binding domain, J. Biol. Chem., 265, 2124–2131.
Belousova, E. A., Vasil’eva, I. A., Moor, N. A., Zatsepin, T. S., Oretskaya, T. S., and Lavrik, O. I. (2013) Clustered DNA lesions containing 5-formyluracil and AP site: repair via the BER system, PLoS One, 8, e68576, doi: https://doi.org/10.1371/journal.pone.0068576
Strauss, P. R., Beard, W. A., Patterson, T. A., and Wilson, S. H. (1997) Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs—Haldane mechanism, J. Biol. Chem., 272, 1302–1307, doi: https://doi.org/10.1074/jbc.272.2.1302.
Chim, N., Harmston, C. A., Guzman, D. J., and Goulding, C. W. (2013) Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis, BMC Struct. Biol., 13, 23, doi: https://doi.org/10.1186/1472-6807-13-23
Wunderlich, M., and Glockshuber, R. (1993) Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli, Protein Sci., 2, 717–726, doi: https://doi.org/10.1002/pro.5560020503
Haugland, R. P. (2005) The Handbook — A Guide to Fluorescent Probes and Labeling Technologies, 10th Edn., Invitrogen Corp., USA.
Dimitriadis, E. K., Prasad, R., Vaske, M. K., Chen, L., Tomkinson, A. E., Lewis, M. S., and Wilson, S. H. (1998) Thermodynamics of human DNA ligase I trimerization and association with DNA polymerase beta, J. Biol. Chem., 273, 20540–20550, doi: https://doi.org/10.1074/jbc.273.32.20540.
Ranalli, T. A., Tom, S., and Bambara, R. A. (2002) AP endonuclease 1 coordinates flap endonuclease 1 and DNA ligase I activity in long patch base excision repair, J. Biol. Chem., 277, 41715–41724, doi: https://doi.org/10.1074/jbc.M207207200
Hanssen-Bauer, A., Solvang-Garten, K., Sundheim, O., Pena-Diaz, J., Andersen, S., Slupphaug, G., Krokan, H. E., Wilson, D. M., 3rd, Akbari, M., and Otterlei, M. (2011) XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage, Environ. Mol. Mutagen., 52, 623–635, doi: https://doi.org/10.1002/em.20663.
Lan, L., Nakajima, S., Oohata, Y., Takao, M., Okano, S., Masutani, M., Wilson, S. H., and Yasui, A. (2004) In situ analysis of repair processes for oxidative DNA damage in mammalian cells, Proc. Natl. Acad. Sci. USA, 101, 13738–13743, doi: https://doi.org/10.1073/pnas.0406048101.
Polo, L. M., Xu, Y., Hornyak, P., Garces, F., Zeng, Z., Hailstone, R., Matthews, S. J., Caldecott, K. W., Oliver, A. W., and Pearl, L. H. (2019) Efficient single-strand break repair requires binding to both poly(ADP-ribose) and DNA by the central BRCT domain of XRCC1, Cell Rep., 26, 573–581, doi: https://doi.org/10.1016/j.celrep.2018.12.082.
Masson, M., Niedergang, C., Schreiber, V., Muller, S., Menissier-de Murcia, J., and de Murcia, G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage, Mol. Cell. Biol., 18, 3563–3571, doi: https://doi.org/10.1128/mcb.18.6.3563.
Vasil’eva, I. A., Moor, N. A., and Lavrik, O. I. (2019) Role of oxidation of XRCC1 protein in regulation of mammalian DNA repair process, Doklady Biochem. Biophys., 489, 357–361, doi: https://doi.org/10.1134/S1607672919060012.
Sukhanova, M., Khodyreva, S., and Lavrik, O. (2010) Poly(ADP-ribose) polymerase 1 regulates activity of DNA polymerase beta in long patch base excision repair, Mutat. Res., 685, 80–89, doi: https://doi.org/10.1016/j.mrfmmm.2009.08.009
Prasad, R., Lavrik, O. I., Kim, S. J., Kedar, P., Yang, X. P., Vande Berg, B. J., and Wilson, S. H. (2001) DNA polymerase beta-mediated long patch base excision repair. Poly(ADP-ribose) polymerase-1 stimulates strand displacement DNA synthesis, J. Biol. Chem., 276, 32411–32414, doi: https://doi.org/10.1074/jbc.C100292200.
Leppard, J. B., Dong, Z., Mackey, Z. B., and Tomkinson, A. E. (2003) Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-ribose) polymerase 1 in DNA single-strand break repair, Mol. Cell. Biol., 23, 5919–5927, doi: https://doi.org/10.1128/mcb.23.16.5919-5927.2003.
Acknowledgements
The authors are grateful to Dr. S. H. Wilson (National Institutes of Health, North Carolina, USA), Dr. J. P. Radicella (UMR217 CNRS/ CEA, France), and Dr. G. Dianov (Oxford University, UK) for kindly providing plasmid vectors for protein expression and to the student Zhao Mingxing of the Novosibirsk State University for participating in the fluorescent titration experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Russian Text © The Author(s), 2020, published in Biokhimiya, 2020, Vol. 85, No. 3, pp. 335–347.
Funding
This work was supported by the Program for Basic Research of the State Academies of Sciences 2013–2020 (project AAAA-A17-117020210022-4; preparation of recombinant proteins) and Russian Science Foundation (project 19-14-00107; study of protein—protein interactions).
Conflict of interest
The authors declare no conflict of interest.
Compliance with ethical norms
The present article does not contain description of studies involving human or animal subjects performed by any of the authors.
Electronic supplementary material
10541_2020_902_MOESM1_ESM.pdf
Effect of Human XRCC1 Protein Oxidation on the Functional Activity of Its Complexes with the Key Enzymes of DNA Base Excision Repair
Rights and permissions
About this article
Cite this article
Vasil’eva, I.A., Moor, N.A. & Lavrik, O.I. Effect of Human XRCC1 Protein Oxidation on the Functional Activity of Its Complexes with the Key Enzymes of DNA Base Excision Repair. Biochemistry Moscow 85, 288–299 (2020). https://doi.org/10.1134/S0006297920030049
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920030049