Abstract
Translocation of conspecific individuals to reduce extinction risk of small, isolated populations and prevent genetic depletion is a powerful tool in conservation biology. An important question is how the translocated individuals influence the long-term genetic composition of the recipient population. Here, we experimentally reinforced a house sparrow (Passer domesticus) population, and examined the impact of this translocation on allele frequencies, levels of heterozygosity and genetic differentiation over six cohorts. We found no permanent increase in the mean number of alleles across loci or levels of observed heterozygosity, but a few alleles private to the translocated individuals remained in the population and we found a short-term increase in heterozygosity. Consequently, genetic differentiation of the recipient population compared to the genetic composition prior to reinforcement was small. The limited genetic impact was due to combined effects of a small probability of establishment and low mating success for the translocated individuals, together with increased genetic drift in the recipient population. Our findings emphasize the importance of selection and genetic drift as forces that may decrease the genetic contribution of reinforcement, especially in small populations. Conservation managers should aim to improve habitat quality in the recipient population to reduce genetic drift following translocation and thereby avoid the need for continued reinforcement. Furthermore, by facilitating establishment success and selecting individuals expected to have high mating success, possibly indicated by sexually selected traits, genetic contribution of released individuals is increased which in turn will decrease reproductive skew and genetic drift.
Similar content being viewed by others
Introduction
Human-induced habitat destruction and over-harvesting are important causes for population declines and threats to biodiversity (Clavero and García-Berthou 2005; Haddad et al. 2015). The persistence of small populations depend on the deterministic growth rate, and their susceptibility to demographic and environmental stochasticity (Lande et al. 2003). Furthermore, small and isolated populations are strongly influenced by genetic drift (Wright 1931; Frankham et al. 2011) and increased levels of inbreeding (Keller and Waller 2002), which may decrease levels of heterozygosity, reduce fitness (Charlesworth and Charlesworth 1999), and ultimately increase extinction risk (Saccheri et al. 1998; Willi et al. 2006).
Because mutation rates are generally low in vertebrates (Lynch et al. 2016), immigration (gene flow) is the main process counteracting the negative impacts of genetic drift and inbreeding (Allendorf and Luikart 2007). Immigration may introduce novel alleles (Hansson et al. 2000; Keller et al. 2001), increase genetic variation (Willi et al. 2006), and decrease levels of inbreeding in receiver populations (Keller et al. 2001; Vilà et al. 2003).
Translocation of conspecific individuals into small and isolated populations (i.e. reinforcement) has demonstrated to be a useful tool for conservation (Griffith et al. 1989; IUCN/SSC 2014). Reinforcements may result in increased growth and recovery of recipient populations (Westemeier et al. 1998; Madsen et al. 1999, 2004). Moreover, reinforcements may also improve the genetic composition by introducing novel alleles, i.e. genetic rescue (Tallmon et al. 2004; Whiteley et al. 2015). For example, in bighorn sheep (Ovis canadensis), reinforcement had positive effects on reproduction and survival, leading to increased allelic diversity and heterozygosity (Hogg et al. 2006). Similarly, survival and heterozygosity increased following reinforcement in Florida panther (Puma concolor coryi) (Johnson et al. 2010). However, such examples of genetic rescue from reinforcement in conservation are still few (Frankham et al. 2014; Frankham 2015). Therefore, experimentally derived information complementing knowledge from conservation is important for our understanding of long-term genetic consequences of reinforcements.
Here we examined multi-generational genetic consequences of a large-scale reinforcement in a house sparrow (Passer domesticus) population. In the first breeding season following the reinforcement, establishment rates and male mating success were lower for translocated individuals than for residents, and individuals mated assortatively based on origin (Ranke et al. 2017). We quantified changes in several genetic parameters: mean number of alleles across loci, proportion of private alleles originating from the translocated and resident individuals, heterozygosity, and pairwise genetic differentiation, FST (proportion of genetic diversity due to allele frequency differences among populations; Holsinger and Weir 2009), between the admixed cohorts and respectively the translocated and resident individuals. The temporal change in FST was also compared to simulated values expected from random genetic drift and demographic stochasticity. In each cohort in the simulation, demographic parameters such as recruit production and survival were acquired from pedigree and ecological data, including the production of recruits based on parental origin in the first breeding season. We predicted that: (1) translocation of genetically unrelated individuals with novel alleles would result in an immediate increase in the mean number of alleles across loci in the recipient population; (2) breeding between translocated and resident individuals would result in an increase in the levels of heterozygosity. However, assortative mating based on origin may delay such an increase in heterozygosity. Finally, if the genetic contribution of translocated and residents is equal, we predict that (3) the admixed cohorts will be equally differentiated from both translocated and resident individuals based on pairwise FST.
Materials and methods
Study area
In 2002, we performed a large-scale experimental reinforcement in a house sparrow population. Individuals were translocated from a source population situated on the island Vega, 12 km off the coast of northern Norway (140 km2, 65.65° N, 11.90° E, Fig. S1), and released into a recipient population inhabiting a similar habitat on the three main islands in the Vikna archipelago (Inner-Vikna, Mellom-Vikna and Ytter-Vikna, 360 km2, 64.91° N, 10.90° E; henceforth referred to as Vikna) located about 95 km south of Vega, and closer to the mainland (ca. 1 km, Fig. S1). We examined genetic consequences of the reinforcement in terms of allelic diversity, levels of heterozygosity and genetic differentiation, in the admixed population and in six subsequent cohorts (i.e. hatched 2002–2007). The house sparrow has low dispersal rates (juveniles < 10% and adults < 2‰; Altwegg et al. 2000) and disperse only short distances (see also, Tufto et al. 2005; Finnish data: 90% < 16 km, Kekkonen et al. 2011), consistent with genetic patterns (Kekkonen et al. 2011; Jensen et al. 2013). In both source and recipient populations, house sparrows live in small colonies on dairy farms (hereafter denoted “subpopulations”, Fig. S1). The subpopulations are separated by at least 1 km and are genetically differentiated (Table S1), despite being interconnected by dispersal, albeit at a low rate (Skjelseth et al. 2007). Some subpopulations have become extinct during the study period (Table 1), and a few have been (re-)colonized after the current study (own personal observations). In addition to the focal subpopulations, sparrows were captured and observed annually in six adjacent populations (within < 5 km) to detect potential dispersal both into and out of the study system.
Experimental design
We describe the protocol prior to the translocation, in the (1) source and (2) recipient populations respectively, (3) the translocation and release of individuals in the recipient population, and (4) monitoring of the admixed population following translocation.
Source population (Vega)
In late February and early March 2002, before onset of breeding, we captured virtually all (~ 90%) house sparrows using mist nets, in the source population on Vega (n = 246 individuals; Table 1; Fig. S1). Birds were marked with an aluminium ring with a unique number and coloured plastic leg-rings. We collected a small blood sample (25 µL) from the brachial vein, which we stored in pure ethanol in a − 25 °C freezer until genotyped (for genotyping procedure, see Online Appendix A). After handling, we released the birds into a large aviary (an inactive and sealed farm-building) in which the birds were kept warm and dry, and supplied with food and water ad libitum. Among the captive birds, 93 individuals were returned to their original colonies on Vega, as part of a selection experiment on tarsus length (Kvalnes et al. 2017). Another 126 individuals were used in the translocation from Vega to the recipient population on Vikna.
Note that between individuals translocated to Vikna and resident individuals on Vikna, neither tarsus length nor body mass (a good predictor of body size; Araya-Ajoy et al. 2019) differed between translocated and resident individuals (Ranke et al. 2017).
Recipient population (Vikna)
In early March 2002, we performed a similar capture session (see methods above) in the recipient population on Vikna. In total 192 individuals were captured in 17 different subpopulations (Table 1; Fig. S1) and transferred to an aviary. Among the birds in the aviary, between 50 and 80% of the individuals of each sex from each subpopulation were randomly selected and then returned to their original subpopulation (n = 120 individuals; Table S2) the 15 March. The remaining individuals (n = 72) were transported by car and released in a suitable habitat on the mainland ca. 110 km southeast of the study area. The removal of individuals mimicked a severe population decline and reduced density regulating effects on population growth.
Translocation and release in the recipient population (Vikna)
The 15–17 March, the source individuals were transported by car in cages from Vega to Vikna. The translocated individuals were allocated to 17 subpopulations in the recipient population so that ratios of origin and sex were equal in each subpopulation (Table S2). To account for an expected loss of individuals commonly associated with translocations (Roe et al. 2010; Germain et al. 2017), the population size of each recipient subpopulation was increased by on average 3.2 individuals (Table S2).
Post-translocation monitoring
In all years from 2002 to 2008, the admixed population was monitored annually during the breeding season from May (from mid-April in 2002) to August, and for 2 weeks during autumn (within the period September–November). Individuals were captured using mist nets and any bird not previously ringed was marked. During each breeding season, nests were located. At 7–13 days after hatching, nestlings were ringed and a blood sample collected (as described for adults above). Annual adult population size was estimated as the total number of adult individuals captured or observed each year (see Table 1). Furthermore, individuals not recorded in year t, but before and after year t, were considered to be present also in year t.
During the first breeding season following reinforcement (2002), 16 unringed individuals were captured in the admixed population. Due to the very low dispersal rate in adult house sparrows (Pärn et al. 2009), these individuals were defined as residents. Thus, the total population size of the admixed population at the time of release was 262 individuals; 126 translocated and 136 residents (Table 1). Out of these, 163 individuals (64 translocated and 99 residents) were still present after one month or later following the release, and were considered established (Table 1).
Genetic analyses
In the genetic analyses for 2002, we included all individuals present at the time of release, i.e. translocated and resident individuals released on Vikna, and additional adults recorded in the recipient population that year. We therefore assume > 90% the individuals present in the admixed population were marked and genetically sampled. In the subsequent years (2003–2008), we included only new recruits (i.e. birds that hatched 2002–2007 and survived to the next year). This was done to avoid influences of overlapping generations and adult age-structure on the temporal genetic dynamics. Note that analyses including all individuals present, instead of only new recruits, gave similar results (Fig. S2), which indicates that our results from the cohorts reflect population wide effects. The cohort year of recruits refers to their hatch year. Within each subpopulation and cohort, very few siblings were detected, reducing potential effects of restrictive breeding. Individuals had to be successfully genotyped at minimum 11 microsatellite loci (of 13 loci in total) to be included. Six individuals were excluded from the genetic analyses (four individuals were unsuccessfully genotyped, and two were incompletely genotyped). An analysis of population genetic structure found a strong correspondence between results based on the same set of neutral microsatellite markers we use in this study and 10 K Single Nucleotide Polymorphisms (SNPs) distributed across the house sparrow genome (Hagen et al. 2013). This suggests that variation in these microsatellite loci reflects genome-wide genetic variation. BLAST searches showed that none of the microsatellites were linked (Table S3).
Statistical analyses
We performed an analysis using STRUCTURE (Pritchard et al. 2000), STRUCTURE HARVESTER (Earl and vonHoldt 2012) and the ‘Evanno method’ (Evanno et al. 2005) to determine the number of genetic clusters prior to reinforcement (Online Appendix B). Then we examined the admixed population at the time of release (both residents and translocated individuals separately and pooled together) and at the time of establishment. Thereafter, we examined the same genetic metrics in all subsequent cohorts. We examined the number of different alleles within and across microsatellite loci using HP-Rare 1.0 (Kalinowski 2005). In the main text we present results based on mean numbers of different alleles, but to account for any potential variation in detection rates between years, we also calculated the allelic richness using rarefaction (i.e. allelic richness across equal population samples; Fig. S2). For individuals considered established and for each subsequent recruit cohort, we quantified alleles private to either the translocated or resident individuals (referred to as “private” also in subsequent cohorts) by first calculating the proportions of each of these two types of alleles within loci and then the mean proportion private alleles across all loci. We assessed differences in the mean number of alleles using paired t-tests, and differences between the mean proportions of private alleles across loci with z-scores predicted from Wilcoxon signed rank tests. To examine temporal trends in the proportions of private alleles present per cohort we used Spearman’s rank correlation (r). We examined the average expected and observed mean heterozygosity across loci using the HIERFSTAT-package in R (Goudet 2005), and assessed differences in mean heterozygosity levels with z-scores predicted from Wilcox signed rank test due to non-normality. Finally, we examined genetic differentiation between the admixed population and all individuals when released, and between established individuals, the subsequent cohorts and (1) translocated individuals from Vega, (2) resident individuals still present at Vikna (see Table 1), based on pairwise FST from HIERFSTAT (Goudet 2005). Significant genetic differentiation was based on 95% confidence intervals not overlapping zero. To examine potential population stratification, deviations from Hardy Weinberg Equilibrium (HWE) were examined using the R-package ‘genepop’ (Rousset 2008; see Table S4).
Simulation
To evaluate the expected influence of genetic drift and demographic stochasticity, we used an individual-based model to simulate allele frequencies in the recipient population in six recruiting cohorts, starting with the established translocated and resident individuals. We assumed equal rates of adult survival of the two groups (see Ranke et al. 2017) and no population structure within the recipient population. The distribution of recruits produced in the first breeding season among parental origin and sex corresponds to the distribution of parent origin and sex following the pedigree (Ranke et al. 2017). All individuals carried their actual microsatellite genotype in year t (year of reinforcement). We obtained annual mean number of recruits produced (\({\uplambda }_{t}\)) and survival rate (\({p}_{t}\)) from empirical data. First, gametes from females (\({f}_{t}\)) and males (\({m}_{t}\)) were obtained by randomly selecting one allele from each locus, from each genotype of potential parents in year t. The number of gametes from each female and male respectively was given by the individual recruit production rate (\({r}_{t}\) ~ Poisson (\({\lambda }_{t}\))). For each cohort, the genotypes of individuals present in year t + 1 were acquired by randomly combining male and female haplotypes present in year t (i.e. to “zygotes” equal to the number of recruits produced in year t + 1), and adding the genotypes of surviving individuals from year t to year t + 1 based on the adult survival rate (\({s}_{t}\) ~ Bernoulli (\({p}_{t}\))). We calculated 95% CI of pairwise FST-values for each post-reinforcement cohort, based on 1000 simulations. Statistical analyses and simulations were performed in R (R Core Team 2018). Whenever needed, significance levels were adjusted to avoid false discoveries (Benjamini and Hochberg 1995).
Results
Genetic changes in the year of reinforcement
The STRUCTURE (Pritchard et al. 2000) and STRUCTURE HARVESTER (Earl and vonHoldt 2012) analysis of source and recipient populations prior to reinforcement inferred two distinct genetic clusters (Online Appendix B: Fig. B1), with a low, but significant level of genetic differentiation.
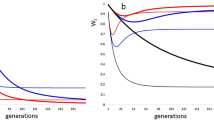
Mean number of alleles was (a) lower for translocated individuals than for residents (translocated ± SE: 10.54 ± 0.96; residents ± SE: 12.08 ± 0.89; t12 = − 6.32, P < 0.001), and (b) higher in the admixed population at time of release than in the recipient population prior to reinforcement (admixed ± SE: 12.54 ± 1.00; resident ± SE: 12.08 ± 0.89; t12 = − 2.52, P = 0.027), and (c) did not differ between the admixed population at time of establishment and the recipient population prior to reinforcement (admixed establishment ± SE: 12.31 ± 0.97; resident ± SE: 12.08 ± 0.89; t12 = − 1.00, P = 0.337; Fig. 1).
Mean number of alleles (± standard errors) across 13 microsatellite loci in an admixed house sparrow population following reinforcement. The left part of the figure shows translocated (triangles), and resident (squares) individuals, and the admixed population (circles) at release (Release), and when established (Establ.) in 2002. The right part shows each subsequent cohort (Y1–Y6, hatched 2002–2007) after reinforcement (circles). Mean number of alleles across loci in the recipient population prior to reinforcement is shown as a dashed horizontal line with ± standard errors as dotted lines
Of 163 different alleles distributed across the 13 microsatellite loci, present in the admixed population at release (Table S5), only six alleles (distributed over five loci) were private to translocated individuals, whereas 26 alleles (distributed over all 13 loci) were private to residents. From release to establishment, one private allele from the translocated individuals and two from the residents were lost.
The mean proportion of alleles private to translocated individuals was lower than those originating from resident individuals at release (translocated ± SE: 0.008 ± 0.004; residents ± SE: 0.020 ± 0.004; z = 2.10, P = 0.036; Fig. 2) and establishment (translocated ± SE: 0.007 ± 0.004; residents ± SE: 0.024 ± 0.005; z = 2.24, P = 0.025; Fig. 2). These differences did not remain significant after accounting for multiple tests.
Mean proportion of private alleles (± standard errors) within each locus in an admixed population of house sparrows following a reinforcement (translocated: n = 5 loci with private alleles; resident: n = 13 loci hosting private alleles). The left part of the figure shows mean proportion of alleles in loci for individuals in the admixed population that were private to translocated (triangles) and resident (squares) individuals at release (Release), and when established (Establ.) in 2002. The right part shows the mean proportion of private alleles from each of the two origins in each subsequent cohort after reinforcement (Y1–Y6, hatched 2002–2007). Mean proportion across loci (n = 5 loci with novel alleles) of alleles that were novel/immigrant alleles (diamonds) is also presented for each of the cohorts after reinforcement. Mean proportion of private alleles in the recipient population prior to reinforcement is shown as a dashed horizontal line with ± standard errors as dotted lines
Heterozygosity did not differ between (a) translocated and resident individuals prior to reinforcement, (b) the admixed population and translocated individuals, and (c) the admixed population and residents, neither at release nor at establishment (all comparisons: z ≤ 0.76, P ≥ 0.449; Fig. 3).
Levels of heterozygosity (± standard errors) in an admixed population of house sparrows following reinforcement. The left part of the figure shows expected (black) and observed (grey) heterozygosity in translocated (triangles), resident (squares), and individuals from both origins pooled in the admixed population (circles) at the time of release (Release), and when established (Establ.) in 2002. The right part shows expected (black) and observed (grey) heterozygosity in each subsequent admixed cohort (circles) after reinforcement, Y1–Y6. Observed heterozygosity in the recipient population prior to reinforcement is shown as a dashed horizontal line with ± standard errors as dotted lines
At release, observed and expected heterozygosity did not differ in translocated (z = − 0.31, P = 0.754) or resident individuals (z = − 1.00, P = 0.319) when analysed separately. However, when admixed, observed heterozygosity was significantly lower than expected heterozygosity at time of release (z = − 2.81, P = 0.005), but not at establishment (z = − 0.75, P = 0.451; Fig. 3).
Finally, prior to reinforcement, translocated and resident individuals were significantly differentiated genetically (pairwise FST = 0.0176, 95% CI 0.0135–0.0219; Fig. 4). At release, the admixed population did not differ significantly from translocated or resident individuals, respectively. However, at establishment, the admixed population was significantly differentiated from the established translocated individuals (FST = 0.0042, 95% CI 0.0020–0.0069; Fig. 4), but not from the established resident individuals (FST = 0.0000, 95% CI − 0.0010−0.0011; Fig. 4).
Genetic differentiation estimated as pairwise FST (with 95% confidence intervals) in an admixed population of house sparrows following reinforcement. The left part of the figure shows FST between the admixed population and the translocated (triangles) and resident individuals (squares), respectively, at the time of release (Release) and when established (Establ.) in 2002. The right part shows pairwise FST between each subsequent admixed cohort (Y1–Y6) and the established translocated individuals (triangles) and established resident individuals from the recipient population (squares), respectively. The horizontal dashed lines represent the pairwise FST estimated between the translocated and resident individuals prior to reinforcement (FST = 0.0176), and the horizontal dotted lines represent the 95% confidence interval (CI 0.0135–0.0219). FST = 0 (i.e. recipient population prior to reinforcement compared with itself) is also shown as a dashed horizontal line. The shaded areas show results from simulations representing expectations under random genetic drift: 95% CI of expected pairwise FST between the admixed cohorts and (1) translocated individuals (light grey), and (2) recipient population (dark grey) at the time of establishment, respectively
Genetic changes in post-reinforcement cohorts
The mean number of alleles across loci of the last cohort of the study (Y6) was significantly lower than for the recipient population immediately after population reduction, prior to reinforcement (t12 = − 6.68, P < 0.001; Fig. 1), the admixed population at release (t12 = − 6.38, P < 0.001) and at establishment (t12 = − 5.90, P < 0.001).
The mean proportion of alleles private to residents was significantly larger than for translocated individuals in all cohorts except one (Y1–Y4 and Y6: z ≥ 2.61, P ≤ 0.009; cohort Y5: z = 1.79, P = 0.073; Fig. 2). Differences in cohort Y2–Y4 remained significant after correcting for multiple tests. There was no significant overall temporal trend in the proportion alleles private to translocated (r = 0.14, P = 0.803) or resident individuals (r = − 0.37, P = 0.497; Fig. 2), and private alleles from both origins were still present in the last cohort (Fig. 2; translocated: three alleles at three loci, mean proportion across loci 0.004 ± 0.002; residents: 13 alleles at nine loci, mean proportion across loci 0.031 ± 0.008).
Six alleles at five different loci (mean proportions across loci and cohorts: ≤ 0.002 ± 0.001, Fig. 4) did not originate from translocated or resident individuals. At least four of these novel alleles were present in adjacent subpopulations outside our study area, and thus likely introduced by natural immigration. Fifteen immigrants were captured during the study period, five originating > 20 km away and 10 from adjacent populations (< 5 km away; Table S6). Unexpectedly, none of the known immigrants carried either of these novel alleles.
In the first cohort of recruits (Y1), observed heterozygosity was significantly lower than expected heterozygosity (z = − 2.01, P = 0.045; Fig. 3). Observed heterozygosity increased significantly between Y1 and Y2 cohort (z = 1.99, P = 0.048; Fig. 3). However, these differences were not significant after correcting for multiple tests. In the following cohorts, heterozygosity decreased to a level similar to pre-reinforcement; thus, it did not differ significantly from the recipient population prior to reinforcement (z = 0.22, P = 0.824).
Finally, we investigated genetic differentiation between each cohort in the admixed population (2002–2007) and (1) translocated and (2) resident individuals at the time of establishment respectively. All cohorts of the admixed population were significantly differentiated from the established translocated individuals (all comparisons: FST ≥ 0.0042, 95% CI ≥ 0.0020–0.0069; Fig. 4). Interestingly, using results from our simulation, the differentiation between established translocated individuals and recruits from the three first cohorts (Y1–Y3) was larger than expected from random genetic drift and demographic stochasticity (Fig. 4). In contrast, the genetic differentiation between established resident individuals and five subsequent cohorts was low and not significantly different from zero (all comparisons: FST ≤ 0.0021, 95% CI’s ≤ − 0.0001–0.0056; Fig. 4). Only cohort Y6 differed significantly from the established resident individuals (FST = 0.0048, 95% CI 0.0008–0.0087; Fig. 4).
Discussion
Genetic changes in the year of reinforcement
Prior to reinforcement, genetic differentiation between the source and recipient populations was low but significant (Online Appendix B: Fig. B1; Fig. 4). Only two individuals were recorded to disperse between the source and recipient population during the study period. Such low rate of exchange, but at the same time low rate of differentiation between the two populations, may indicate immigration from genetically similar populations (Kekkonen et al. 2011; Jensen et al. 2013). Thus, the small genetic differences between the source and resident populations (Fig. 4; Online Appendix B) are likely a result of genetic drift counteracted by low levels of gene flow (Holand et al. 2011).
Low allelic variation and few private alleles among translocated individuals suggest that the source population may not have been sufficiently genetically variable and differentiated to increase allelic diversity of the recipient population. In fact, this may reflect the constraints often faced by managers: genetic information prior to translocation is limited and suitable source populations are scarce (Heber et al. 2013). However, monitoring the trajectories of private alleles from both origins should provide a representative evaluation of genetic contribution from translocated individuals, also into genetically diverse populations. Though, it provides no information regarding the introgression of individuals carrying these alleles.
At release, the admixed population were equally differentiated from both the source and recipient population. This was expected, given that equal numbers of translocated and resident individuals were released. However, already at the time of establishment, the admixed population was more differentiated from the translocated individuals than from the resident individuals (Fig. 4). This is likely due to the lower establishment success of translocated individuals compared to residents found in this experiment (50.8% vs. 71.3%; Table 1; Ranke et al. 2017), and in other translocations (Roe et al. 2010; Germain et al. 2017). Translocated individuals often move over larger distances experiencing higher mortality following translocations (Roe et al. 2010). Low establishment success may at an early stage thus contribute to a long-term limited genetic impact of reinforcements.
Genetic changes in the post-reinforcement cohorts
There was a temporal decrease in mean number of alleles in the admixed population (Fig. 1), opposite of the intended outcome of reinforcements (Tallmon et al. 2004; Whiteley et al. 2015). The decrease in mean number of alleles across all cohorts was likely caused by genetic drift resulting from an overall decline in population size (Engen et al. 2007; Table 1). This highlights the importance of mitigating the initial conditions leading to population decline, in order to avoid sustained negative influences on population size and a corresponding increased genetic drift (Bouzat et al. 2009; Heber et al. 2013; Bateson et al. 2014). Thus, to decrease the dependence of repeated reinforcements, improvement of habitat quality would be needed (Frankham 2015).
In the admixed population, the mean proportion of alleles private to resident individuals was higher than for translocated individuals in three cohorts. This suggests a larger genetic contribution from resident individuals, at least in some cohorts (Fig. 2). In this experiment, the source population was more isolated than the recipient population based on the different distance to mainland, which may have contributed to the lower allelic diversity in the source population. Moreover, higher establishment rate and mating success further contributed to the larger proportion of private alleles originating from the recipient population (Fig. 2). This difference in genetic contribution is apparent already after the first breeding season (Online Appendix C). Six novel alleles were detected during the study period, indicating some immigration from adjacent populations where at least four of these alleles were present (Fig. 2). None of these naturally introduced alleles were present for more than two consecutive cohorts. This contrasts other more inbred recipient populations, where immigrant alleles may have a large impact on population genetic diversity (Åkesson et al. 2016), especially after recent bottlenecks (Keller et al. 2001). Natural immigrants into the recipient population might have been subject to lower establishment rates and mating success, similarly as the translocated individuals. Moreover, these novel alleles were also present in low numbers, and were therefore more vulnerable to be lost due to genetic drift.
In the first cohort following the reinforcement, there was no increase in observed heterozygosity, but instead lower observed heterozygosity than expected (Fig. 3). Fewer heterozygotes is expected according to the Wahlund effect, i.e. lower observed than expected heterozygosity in subdivided populations (Wahlund 1928). Genetic mixing between the two origins were low likely due to assortative mating based on origin i.e. recruits in the first cohort were mostly offspring of translocated-translocated pairs or resident-resident pairs (Ranke et al. 2017). In the second cohort, these restrictions seem to disappear (Fig. 3), increasing the observed heterozygosity, suggesting that the delayed increase in observed heterozygosity by one cohort was due to limited mixing of the two origins following the translocation. In contrast to our result, other studies have demonstrated more immediate increase in heterozygosity already in the first cohort following reinforcement (Yamamoto et al. 2006; Heber et al. 2013). For example, even when using inbred source populations, heterozygosity increased following reinforcements into two inbred populations of South Island robin (Petroica australis) (Heber et al. 2013). In our experiment, the increase in observed heterozygosity was only temporary. During the four subsequent cohorts, it returned to a pre-reinforcement level (Fig. 3), likely due to the high levels of heterozygosity in the recipient population prior to reinforcement. In contrast, for severely inbred populations, reinforcement may lead to long-term increase in heterozygosity, as found in e.g. the Florida panther (Johnson et al. 2010).
Temporal changes in genetic differentiation are common in reinforced populations (reviewed in Champagnon et al. 2012), due to the influence from genetically distinct individuals (Frankham 2015). However, in the focal reinforced population, only the last cohort showed genetic differentiation when compared to the recipient population prior to reinforcement (Fig. 4). Populations unaffected by reinforcement often show such small temporal change in allele frequencies (Tessier and Bernatchez 1999), provided that populations are sufficiently large and genetic drift negligible. In contrast, all cohorts were genetically differentiated from the translocated individuals (Fig. 4). The genetic differentiation was actually more rapid than expected from random genetic drift and demographic stochasticity according to our simulation, even when accounting for the assortative mating present among the parental individuals (Fig. 4). Together, these findings suggest small genetic contributions from translocated individuals to the recipient population. In contrast, reinforcement of isolated and genetically depleted recipient populations may have large impact on genetic differentiation. For instance, in white-spotted charr (Salvelinus leucomaenis) genetic differentiation between source and recipient populations had disappeared already in the cohort following the reinforcement (Yamamoto et al. 2006).
Implications for conservation
Our results suggest that interactions between translocated and resident individuals following reinforcement may introduce additional challenges compared to reintroductions (see Ewen et al. 2012). This study underlines the importance of detailed temporal genetic assessment following reinforcement of populations, monitoring the allelic diversity, unique alleles, heterozygosity and genetic differentiation. Thorough genetic analyses on several time steps were needed to identify the underlying reasons for the limited genetic effects of reinforcement. In our study, both establishment and mating success were lower for translocated individuals than residents. We reveal how these important ecological and behavioural factors might impede genetic contribution of reinforcement in threatened populations. Any fitness disadvantage for translocated individuals may reduce their long-term genetic impact on the recipient population. Moreover, during this study, several farms in the recipient population ceased operation i.e. equivalent to habitat destruction for the house sparrow. This resulted in local population extinctions (Table 1), reduced population size and increased fragmentation, which in turn increased genetic drift and loss of genetic diversity. Therefore, any threats to the resident population should be reduced prior to reinforcement. Otherwise, sustained negative impacts on population size and allelic diversity will lead to the population being dependent on repeated reinforcements (Bouzat et al. 2009; Frankham 2015). Our findings highlight the importance of careful genetic examination of both source and recipient populations prior to reinforcement, and we stress the importance of implementing guidelines for translocations (Bateson et al. 2014; Frankham et al. 2014; IUCN/SSC 2014). Still, our experiment reveals that even when allelic diversity of the source population is low relative to the recipient population, reinforcement may introduce novel alleles and cause a short-term increase in heterozygosity.
Data availability
All data used in this study are made available in Dryad Digital Repository: https://doi.org/10.5061/dryad.612jm640w (Ranke et al. 2020).
References
Åkesson M, Liberg O, Sand H, Wabakken P, Bensch S, Flagstad Ø (2016) Genetic rescue in a severely inbred wolf population. Mol Ecol 25:4745–4756
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell Publishing, Malden
Altwegg R, Ringsby TH, Sæther B-E (2000) Phenotypic correlates and consequences of dispersal in a metapopulation of house sparrows Passer domesticus. J Anim Ecol 69:762–770
Araya-Ajoy Y, Ranke PS, Kvalnes T et al (2019) Characterizing morphological (co)variation using structural equation models: body size, allometric relationships and evolvability in a house sparrow metapopulation. Evolution 73:452–466
Bateson ZW, Dunn PO, Hull SD, Henschen AE, Johnson JA, Whittingham LA (2014) Genetic restoration of a threatened population of greater prairie-chickens. Biol Conserv 174:12–19
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Bouzat JL, Johnson JA, Toepfer JE, Simpson SA, Esker TL, Westemeier RL (2009) Beyond the beneficial effects of translocations as an effective tool for genetic restoration of isolated populations. Conserv Genet 10:191–201
Champagnon J, Elmberg J, Guillemain M, Gauthier-Clerc M, Lebreton J-D (2012) Conspecifics can be aliens too: a review of effects of restocking practices in vertebrates. J Nat Conserv 20:231–241
Charlesworth B, Charlesworth D (1999) The genetic basis of inbreeding depression. Genet Res 74:329–340
Clavero M, García-Berthou E (2005) Invasive species are a leading cause of animal extinctions. Trends Ecol Evol 20:110
Earl D, vonHoldt B (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res 4:359–361
Engen S, Ringsby TH, Sæther B-E, Lande R, Jensen H, Lillegård M, Ellegren H (2007) Effective size of fluctuating populations with two sexes and overlapping generations. Evolution 61:1873–1885
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewen JG, Armstrong DP, Parker KA, Seddon PJ (eds) (2012) Reintroduction biology: integrating science and management. Wiley-Blackwell, West Sussex
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618
Frankham R, Ballou JD, Briscoe DA (2011) Introduction to conservation genetics, 2nd edn. Cambridge University Press, Cambridge
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Germain M, Pärt T, Doligez B (2017) Lower settlement following a forced displacement experiment: nonbreeding as a dispersal cost in a wild bird? Anim Behav 133:109e
Goudet J (2005) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186
Griffith B, Scott JM, Carpenter JW, Reed C (1989) Translocation as a species conservation tool: status and strategy. Science 245:477–480
Haddad NM, Brudvig LA, Clobert J et al (2015) Habitat fragmentation and its lasting impact on earth’s ecosystems. Sci Adv 1:e1500052
Hagen IJ, Billing AM, Rønning B, Pedersen SA, Pärn H, Slate J, Jensen H (2013) The easy road to genome-wide medium density SNP screening in a non-model species: development and application of a 10 K SNP-chip for the house sparrow (Passer domesticus). Mol Ecol Resour 13:429–439
Hansson B, Bensch S, Hasselquist D, Lillandt B-G, Wennerberg L, Von Schantz T (2000) Increase of genetic variation over time in a recently founded population of great reed warblers (Acrocephalus arundinaceus) revealed by microsatellites and DNA fingerprinting. Mol Ecol 9:1529–1538
Heber S, Varsani A, Kuhn S, Girg A, Kempenaers B, Briskie J (2013) The genetic rescue of two bottlenecked South Island robin populations using translocations of inbred donors. Proc R Soc B 280:20122228
Hogg JT, Forbes SH, Steele BM, Luikart G (2006) Genetic rescue of an insular population of large mammals. Proc R Soc B 273:1491–1499
Holand AM, Jensen H, Tufto J, Moe R (2011) Does selection or genetic drift explain geographic differentiation of morphological characters in house sparrows Passer domesticus? Genet Res 93:367–379
Holsinger KE, Weir BS (2009) Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet 10:639–650
IUCN/SSC (2014) Guidelines for reintroductions and other conservation translocations. Version 2.0. p. viiii + 57 pp. IUCN Species Survival Commission, Gland
Jensen H, Moe R, Hagen IJ, Holand AM, Kekkonen J, Tufto J, Sæther B-E (2013) Genetic variation and structure of house sparrow populations: is there an island effect? Mol Ecol 22:1792–1805
Johnson WE, Onorato DP, Roelke ME et al (2010) Genetic restoration of the Florida panther. Science 329:1641–1645
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Kekkonen J, Seppä P, Hanski IK, Jensen H, Väisänen RA, Brommer JE (2011) Low genetic differentiation in a sedentary bird: house sparrow population genetics in a contiguous landscape. Heredity 106:183–190
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Keller LF, Jeffery KJ, Arcese P, Beaumont MA, Hochachka WM, Smith JNM, Bruford MW (2001) Immigration and the ephemerality of a natural population bottleneck: evidence from molecular markers. Proc R Soc B 268:1387–1394
Kvalnes T, Ringsby TH, Jensen H et al (2017) Reversal of response to artificial selection on body size in a wild passerine bird. Evolution 71:2062–2079
Lande R, Engen S, Sæther B-E (2003) Stochastic population dynamics in ecology and conservation. Oxford University Press, London
Lynch M, Ackerman MS, Gout J-F, Long H, Sung W, Thomas WK, Foster PL (2016) Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet 17:704–714
Madsen T, Shine R, Olsson M, Wittzell H (1999) Conservation biology: restoration of an inbred adder population. Nature 402:34–35
Madsen T, Ujvari B, Olsson M (2004) Novel genes continue to enhance population growth in adders (Vipera berus). Biol Conserv 120:145–147
Pärn H, Jensen H, Ringsby TH, Sæther B-E (2009) Sex-specific fitness correlates of dispersal in a house sparrow metapopulation. J Anim Ecol 78:1216–1225
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Ranke PS, Skjelseth S, Pärn H et al (2017) Demographic influences of translocated individuals on a resident population of house sparrows. Oikos 126:1410–1418
Ranke PS (2020) Microsatellite data supporting the study of multi-generational genetic consequences of reinforcement in a bird metapopulation. https://doi.org/10.5061/dryad.612jm640w
Roe JH, Frank MR, Gibson SE, Attum O, Kingsbury BA (2010) No place like home: an experimental comparison of reintroduction strategies using snakes. J Appl Ecol 47:1253–1261
Rousset F (2008) GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392:491–494
Skjelseth S, Ringsby TH, Tufto J, Jensen H, Sæther B-E (2007) Dispersal of introduced house sparrows Passer domesticus: an experiment. Proc R Soc B 274:1763–1771
Tallmon DA, Luikart G, Waples RS (2004) The alluring simplicity and complex reality of genetic rescue. Trends Ecol Evol 19:489–496
Tessier N, Bernatchez L (1999) Stability of population structure and genetic diversity across generations assessed by microsatellites among sympatric populations of landlocked Atlantic salmon (Salmo salar L.). Mol Ecol 8:169–179
Tufto J, Ringsby TH, Dhondt AA, Adriaensen F, Matthysen E (2005) A parametric model for estimation of dispersal patterns applied to five passerine spatially structured populations. Am Nat 165:E13–E26
Vilà C, Sundqvist A-K, Flagstad Ø et al (2003) Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc R Soc B 270:91–97
Wahlund S (1928) The constitution of populations and correlation phenomena regarded from a genetic viewpoint. Hereditas 11:65–106
Westemeier RL, Brawn JD, Simpson SA et al (1998) Tracking the long-term decline and recovery of an isolated population. Science 282:1695–1698
Whiteley AR, Fitzpatrick SW, Funk WC, Tallmon DA (2015) Genetic rescue to the rescue. Trends Ecol Evol 30:42–49
Willi Y, Van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Syst 37:433–458
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Yamamoto S, Maekawa K, Tamate T, Koizumi I, Hasegawa K, Kubota H (2006) Genetic evaluation of translocation in artificially isolated populations of white-spotted charr (Salvelinus leucomaenis). Fish Res 78:352–358
Acknowledgements
Open Access funding provided by NTNU Norwegian University of Science and Technology (incl St. Olavs Hospital - Trondheim University Hospital). We thank land owners at Vikna and Vega for kindly allowing us to perform our work at their properties, technicians in field and at the lab, and two anonymous reviewers for improving the manuscript. The research was carried out in accordance with permits from the Norwegian Environment Agency and the Bird Ringing Centre at Stavanger Museum, Norway. This study was funded by the Norwegian Research Council (Project Number 221956, and the Centres of Excellence funding scheme, Project Number 223757) and the Norwegian University of Science and Technology.
Author information
Authors and Affiliations
Contributions
Author contribution statement THR, HJ and B-ES conceived the study. The experiment was carried out by THR and HJ with the help of field workers. Laboratory analyses were carried out by SS, AMB and ÅABP, and was supervised by HJ. Data analyses were carried out by IJH, PSR and SS, with support from HJ and HP. Writing of the manuscript was led by PSR, with input from all co-authors. All authors contributed to manuscript editing and revisions.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ranke, P.S., Skjelseth, S., Hagen, I.J. et al. Multi-generational genetic consequences of reinforcement in a bird metapopulation. Conserv Genet 21, 603–612 (2020). https://doi.org/10.1007/s10592-020-01273-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01273-7