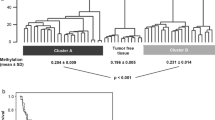
Abstract
Patterns of DNA methylation are significantly altered in cancers. Interpreting the functional consequences of DNA methylation requires the integration of multiple forms of data. The recent advancement in the next-generation sequencing can help to decode this relationship and in biomarker discovery. In this study, we investigated the methylation patterns of papillary renal cell carcinoma (PRCC) and its relationship with the gene expression using The Cancer Genome Atlas (TCGA) multi-omics data. We found that the promoter and body of tumor suppressor genes, microRNAs and gene clusters and families, including cadherins, protocadherins, claudins and collagens, are hypermethylated in PRCC. Hypomethylated genes in PRCC are associated with the immune function. The gene expression of several novel candidate genes, including interleukin receptor IL17RE and immune checkpoint genes HHLA2, SIRPA and HAVCR2, shows a significant correlation with DNA methylation. We also developed machine learning models using features extracted from single and multi-omics data to distinguish early and late stages of PRCC. A comparative study of different feature selection algorithms, predictive models, data integration techniques and representations of methylation data was performed. Integration of both gene expression and DNA methylation features improved the performance of models in distinguishing tumor stages. In summary, our study identifies PRCC driver genes and proposes predictive models based on both DNA methylation and gene expression. These results on PRCC will aid in targeted experiments and provide a strategy to improve the classification accuracy of tumor stages.








Similar content being viewed by others
Data availability
The code for building the models and additional data can be found at https://github.com/NPSDC/IGAMS.
Abbreviations
- BEMKL:
-
Bayesian efficient multiple kernel learning
- ccRCC:
-
Clear cell renal cell carcinoma
- DEGs:
-
Differentially expressed genes
- DMCs:
-
Differentially methylated CpG sites
- GL:
-
Group lasso
- KNN:
-
k-Nearest neighbors
- MKL:
-
Multiple kernel learning
- NB:
-
Naive Bayes
- PRCC:
-
Papillary renal cell carcinoma
- RCC:
-
Renal cell carcinoma
- RF:
-
Random forest
- SC:
-
Shrunken centroids
- SVM:
-
Support vector machine
- TSGs:
-
Tumor suppressor genes
References
Ali M, Khan SA, Wennerberg K, Aittokallio T (2018) Global proteomics profiling improves drug sensitivity prediction: results from a multi-omics, pan-cancer modeling approach. Bioinformatics 34(8):1353–1362
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106
Anderson AC (2014) Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol Res 2(5):393–398
Arai E, Chiku S, Mori T, Gotoh M, Nakagawa T, Fujimoto H, Kanai Y (2012) Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis 33(8):1487–1493
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA (2014) Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30(10):1363–1369
Bartel CA, Parameswaran N, Cipriano R, Jackson MW (2016) FAM83 proteins: fostering new interactions to drive oncogenic signaling and therapeutic resistance. Oncotarget 7(32):52597–52612
Baylin SB (2006) DNA methylation and gene silencing in cancer. ChemInform. https://doi.org/10.1002/chin.200622296
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11(10):726–734
Chen F, Bai J, Li W, Mei P, Liu H, Li L, Pan Z, Wu Y, Zheng J (2013) RUNX3 suppresses migration, invasion and angiogenesis of human renal cell carcinoma. PLoS ONE 8(2):e56241
Chen F, Zhang Y, Şenbabaoğlu Y, Ciriello G, Yang L, Reznik E, Shuch B et al (2016) Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep 14(10):2476–2489
Chen G, Wang Y, Wang L, Xu W (2017) Identifying prognostic biomarkers based on aberrant DNA methylation in kidney renal clear cell carcinoma. Oncotarget 8(3):5268–5280
Chen D, Chen W, Xu Y, Zhu M, Xiao Y, Shen Y, Zhu S, Cao C, Xu X (2019) Upregulated immune checkpoint HHLA2 in clear cell renal cell carcinoma: a novel prognostic biomarker and potential therapeutic target. J Med Genet 56(1):43–49
Chicco D (2017) Ten quick tips for machine learning in computational biology. BioData Mining 10:35
Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS et al (2016) TCGAbiolinks: an R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44(8):e71
Cortes C, Vapnik V (1995) Support-Vector networks. Mach Learn 20(3):273–297
Costello JC, Heiser LM, Georgii E, Gönen M, Menden MP, Wang NJ, Bansal M et al (2014) A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 32(12):1202–1212
De Caceres II, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P (2006) Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res 66(10):5021–5028
Deckers IA, Schouten LJ, Van Neste L, Van Vlodrop IJ, Soetekouw PM, Baldewijns MM, Jeschke J et al (2015) Promoter methylation of CDO1 identifies clear-cell renal cell cancer patients with poor survival outcome. Clin Cancer Res 21(15):3492–3500
Díaz-Uriarte R, De Andres SA (2006) Gene selection and classification of microarray data using random forest. BMC Bioinform 7:3
Durinck S, Stawiski EW, Pavía-Jiménez A, Modrusan Z, Kapur P, Jaiswal BS, Zhang N et al (2015) Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. https://doi.org/10.1038/ng.3146
Eden A, Gaudet F, Waghmare A, Jaenisch R (2003) Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300(5618):455
Ellinger J, Holl D, Nuhn P, Kahl P, Haseke N, Staehler M, Siegert S et al (2011) DNA hypermethylation in papillary renal cell carcinoma. BJU Int 107(4):664–669
Feng ZH, Fang Y, Zhao LY, Lu J, Wang YQ, Chen ZH, Huang Y et al (2017) RIN1 promotes renal cell carcinoma malignancy by activating EGFR signaling through Rab25. Cancer Sci 108(8):1620–1627
Gonen M (2012) bayesian efficient multiple kernel learning. arXiv [cs.LG]. arXiv. http://arxiv.org/abs/1206.6465
Gönen M, Alpaydın E (2011) Multiple kernel learning algorithms. J Mach Learn Res 12:2211–2268
Guan Z, Zhang J, Wang J, Wang H, Zheng F, Peng J, Xu Y et al (2014) SOX1 down-regulates β-catenin and reverses malignant phenotype in nasopharyngeal carcinoma. Mol Cancer 13:257
Guo C, Zhao D, Zhang Q, Liu S, Sun MZ (2018) miR-429 suppresses tumor migration and invasion by targeting CRKL in hepatocellular carcinoma via inhibiting Raf/MEK/ERK pathway and epithelial–mesenchymal transition. Sci Rep 8(1):2375
Hansson J, Lindgren D, Nilsson H, Johansson E, Johansson M, Gustavsson L, Axelson H (2017) Overexpression of functional SLC6A3 in clear cell renal cell carcinoma. Clin Cancer Res 23(8):2105–2115
Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, Flagg K et al (2017) DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci USA 114(28):7414–7419
He C, Zhao X, Jiang H, Zhong Z, Xu R (2015) Demethylation of miR-10b plays a suppressive role in ccRCC cells. Int J Clin Exp Pathol 8(9):10595–10604
He RQ, Qin MJ, Lin P, Luo YH, Ma J, Yang H, Hu XH, Chen G (2018) Prognostic significance of LncRNA PVT1 and its potential target gene network in human cancers: a comprehensive inquiry based upon 21 cancer types and 9972 Cases. Cell Physiol Biochem 46(2):591–608
Hsieh JJ, Le V, Cao D, Cheng EH, Creighton CJ (2018) Genomic classifications of renal cell carcinoma: a critical step towards the future application of personalized kidney cancer care with pan-omics precision. J Pathol 244(5):525–537
Hu Y, Xing J, Wang L, Huang M, Guo X, Chen L, Lin M et al (2011) RGS22, a novel cancer/testis antigen, inhibits epithelial cell invasion and metastasis. Clin Exp Metas 28(6):541–549
Hu CA, Klopfer EI, Ray PE (2012) Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. https://doi.org/10.1016/j.febslet.2012.03.002
Huang S, Chaudhary K, Garmire LX (2017) More is better: recent progress in multi-omics data integration methods. Front Genet 8:84
IlluminaHumanMethylation450kanno, Hansen K (2014) ilmn12. hg19: annotation for illumina’s 450k methylation arrays. R Package Version 0.2. 1
Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA (2012) Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 41(1):200–209
Jiang Y, Shi X, Zhao Q, Krauthammer M, Rothberg BE, Ma S (2016) Integrated analysis of multidimensional omics data on cutaneous melanoma prognosis. Genomics 107(6):223–230
Jiao Y, Widschwendter M, Teschendorff AE (2014) A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics 30(16):2360–2366
Jonasch E, Gao J, Rathmell WK (2014) Renal cell carcinoma. BMJ 349:g4797
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492
Kang MH, Choi H, Oshima M, Cheong JH, Kim S, Lee JH, Park YS et al (2018) Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer. Nat Commun 9(1):1920
Kim D, Joung JG, Sohn KA, Shin H, Park YR, Ritchie MD, Kim JH (2015) Knowledge boosting: a graph-based integration approach with multi-omics data and genomic knowledge for cancer clinical outcome prediction. J Am Med Inform Assoc JAMIA 22(1):109–120
Kim D, Li R, Lucas A, Verma SS, Dudek SM, Ritchie MD (2017) Using knowledge-driven genomic interactions for multi-omics data analysis: metadimensional models for predicting clinical outcomes in ovarian carcinoma. J Am Med Inform Assoc JAMIA 24(3):577–587
Klacz J, Wierzbicki PM, Wronska A, Rybarczyk A, Stanislawowski M, Slebioda T, Olejniczak A, Matuszewski M, Kmiec Z (2016) Decreased expression of rassf1a tumor suppressor gene is associated with worse prognosis in clear cell renal cell carcinoma. Int J Oncol 48(1):55–66
Kluzek K, Bluyssen HA, Wesoly J (2015) The epigenetic landscape of clear-cell renal cell carcinoma. J Kidney Cancer VHL 2(3):90–104
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw377
Kulis M, Heath S, Bibikova M, Queirós AC, Navarro A, Clot G, Martínez-Trillos A et al (2012) Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 44(11):1236–1242
Lasseigne BN, Brooks JD (2018) The role of DNA methylation in renal cell carcinoma. Mol Diagn Ther. https://doi.org/10.1007/s40291-018-0337-9
Lasseigne BN, Burwell TC, Patil MA, Absher DM, Brooks JD, Myers RM (2014) DNA methylation profiling reveals novel diagnostic biomarkers in renal cell carcinoma. BMC Med 12:235
Li X, Chen C, Wang F, Huang W, Liang Z, Xiao Y, Wei K et al (2014) KCTD1 suppresses canonical Wnt signaling pathway by enhancing β-catenin degradation. PLoS ONE 9(4):e94343
Liao R, Sun J, Wu H, Yi Y, Wang JX, He HW, Cai XY et al (2013) High expression of IL-17 and IL-17RE associate with poor prognosis of hepatocellular carcinoma. J Exp Clin Cancer Res 32:3
Lin E, Lane HY (2017) Machine learning and systems genomics approaches for multi-omics data. Biomark Res 5:2
Liu W, Liu H, Liu Y, Xu L, Zhang W, Zhu Y, Xu J, Gu J (2014) Prognostic significance of p21-activated kinase 6 expression in patients with clear cell renal cell carcinoma. Ann Surg Oncol 21(Suppl 4):S575–S583
Liu X, Yao D, Liu C, Cao Y, Yang Q, Sun Z, Liu D (2016a) Overexpression of ABCC3 promotes cell proliferation, drug resistance, and aerobic glycolysis and is associated with poor prognosis in urinary bladder cancer patients. Tumour Biol 37(6):8367–8374
Liu Y, Jin X, Li Y, Ruan Y, Lu Y, Yang M, Lin D et al (2016b) DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J Exp Clin Cancer Res 35(1):120
Llinàs-Arias P, Esteller M (2017) Epigenetic inactivation of tumour suppressor coding and non-coding genes in human cancer: an update. Open Biol. https://doi.org/10.1098/rsob.170152
Lu AG, Feng H, Pu-Xiong-Zhi Wang DP, Han XH, Zheng MH (2012) Emerging roles of the ribonucleotide reductase M2 in colorectal cancer and ultraviolet-induced DNA damage repair. World J Gastroenterol 18(34):4704–4713
Mankoo PK, Shen R, Schultz N, Levine DA, Sander C (2011) Time to recurrence and survival in serous ovarian tumors predicted from integrated genomic profiles. PLoS ONE 6(11):e24709
Matthews BW (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochem Biophys Acta 405(2):442–451
Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE et al (2010) Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466(7303):253–257
McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, Beroukhim R et al (2013) The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell. https://doi.org/10.1016/j.ccr.2013.08.004
McMahon KW, Karunasena E, Ahuja N (2017) The roles of DNA methylation in the stages of cancer. Cancer J. https://doi.org/10.1097/ppo.0000000000000279
Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM (2016) The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol 70(1):93–105
Modi PK, Singer EA (2016) Improving our understanding of papillary renal cell carcinoma with integrative genomic analysis. Ann Transl Med 4(7):143
Morris MR, Latif F (2017) The epigenetic landscape of renal cancer. Nat Rev Nephrol 13(1):47–60
Morris MR, Maher ER (2010) Epigenetics of renal cell carcinoma: the path towards new diagnostics and therapeutics. Genome Med 2(9):59
Morris MR, Gentle D, Abdulrahman M, Maina EN, Gupta K, Banks RE, Wiesener MS et al (2005) Tumor suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma. Cancer Res 65(11):4598–4606
Morrissey C, Martinez A, Zatyka M, Agathanggelou A, Honorio S, Astuti D, Morgan NV et al (2001) Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res 61(19):7277–7281
Nishiwada S, Sho M, Yasuda S, Shimada K, Yamato I, Akahori T, Kinoshita S, Nagai M, Konishi N, Nakajima Y (2015) Nectin-4 expression contributes to tumor proliferation, angiogenesis and patient prognosis in human pancreatic cancer. J Exp Clin Cancer Res 34:30
Paluszczak J, Wiśniewska D, Kostrzewska-Poczekaj M, Kiwerska K, Grénman R, Mielcarek-Kuchta D, Jarmuż-Szymczak M (2017) Prognostic significance of the methylation of wnt pathway antagonists-CXXC4, DACT2, and the inhibitors of sonic hedgehog signaling-ZIC1, ZIC4, and HHIP in head and neck squamous cell carcinomas. Clin Oral Investig 21(5):1777–1788
Quadri HS, Aiken TJ, Allgaeuer M, Moravec R, Altekruse S, Hussain SP, Miettinen MM, Hewitt SM, Rudloff U (2017) Expression of the scaffold connector enhancer of kinase suppressor of Ras 1 (CNKSR1) is correlated with clinical outcome in pancreatic cancer. BMC Cancer 17(1):495
Rahimi A, Gönen M (2018) Discriminating early- and late-stage cancers using multiple kernel learning on gene sets. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty239
Rapetti-Mauss R, Bustos V, Thomas W, McBryan J, Harvey H, Lajczak N, Madden SF et al (2017) Bidirectional KCNQ1:β-catenin interaction drives colorectal cancer cell differentiation. Proc Natl Acad Sci USA 114(16):4159–4164
Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM, Powers S (2013) Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 145(6):1424–1435.e1–25
Ricketts CJ, Morris MR, Gentle D, Brown M, Wake N, Woodward ER, Clarke N, Latif F, Maher ER (2012) Genome-wide CpG island methylation analysis implicates novel genes in the pathogenesis of renal cell carcinoma. Epigenetics 7(3):278–290
Ricketts CJ, De Cubas AA, Fan H, Smith CC, Lang M, Reznik E, Bowlby R et al (2018) The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep 23(12):3698
Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, Fulda S et al (2010) CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut 59(8):1101–1110
Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D (2015) Methods of integrating data to uncover genotype–phenotype interactions. Nat Rev Genet 16(2):85–97
Saha SK, Islam SR, Kwak KS, Rahman MS, Cho SG (2019) PROM1 and PROM2 expression differentially modulates clinical prognosis of cancer: a multiomics analysis. Cancer Gene Ther. https://doi.org/10.1038/s41417-019-0109-7
Saito T, Rehmsmeier M (2015) The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE. https://doi.org/10.1371/journal.pone.0118432
Salama M, Benitez-Riquelme D, Elabd S, Munoz L, Zhang P, Glanemann M, Mione MC et al (2019) Fam83F induces p53 stabilisation and promotes its activity. Cell Death Differ. https://doi.org/10.1038/s41418-019-0281-1
Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M (2011) Validation of a dna methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. https://doi.org/10.4161/epi.6.6.16196
Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, Michaud A et al (2015) Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol Cell 57(5):769–783
Seoane JA, Day IN, Gaunt TR, Campbell C (2014) A pathway-based data integration framework for prediction of disease progression. Bioinformatics 30(6):838–845
Shen S, Wang G, Shi Q, Zhang R, Zhao Y, Wei Y, Chen F, Christiani DC (2017) Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin Epigenet 9:88
Shenoy N, Vallumsetla N, Zou Y, Galeas JN, Shrivastava M, Hu C, Susztak K, Verma A (2015) Role of DNA methylation in renal cell carcinoma. J Hematol Oncol 8:88
Singh NP, Bapi RS, Vinod PK (2018) Machine learning models to predict the progression from early to late stages of papillary renal cell carcinoma. Comput Biol Med 100:92–99
Smutna V, Dieci MV, Lefebvre C, Scott V, Andre F, Fromigue O (2014) Abstract 435: is PYGM dysregulation involved in breast cancer cell metabolism. Cancer Res. https://doi.org/10.1158/1538-7445.am2014-435
Snijders AM, Lee SY, Hang B, Hao W, Bissell MJ, Mao JH (2017) FAM83 family oncogenes are broadly involved in human cancers: an integrative multi-omics approach. Mol Oncol 11(2):167–179
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45(4):427–437
Sorokin AV, Nair BC, Wei Y, Aziz KE, Evdokimova V, Hung MC, Chen J (2015) Aberrant expression of proPTPRN2 in cancer cells confers resistance to apoptosis. Cancer Res 75(9):1846–1858
Soshnikova N, Duboule D (2009) Epigenetic regulation of vertebrate hox genes: a dynamic equilibrium. Epigenetics 4(8):537–540
Stott-Miller M, Zhao S, Wright JL, Kolb S, Bibikova M, Klotzle B, Ostrander EA, Fan JB, Feng Z, Stanford JL (2014) Validation study of genes with hypermethylated promoter regions associated with prostate cancer recurrence. Cancer Epidemiol Biomark Prev 23(7):1331–1339
Su Y, Lin Y, Zhang L, Liu B, Yuan W, Mo X, Wang X et al (2014) CMTM3 inhibits cell migration and invasion and correlates with favorable prognosis in gastric cancer. Cancer Sci 105(1):26–34
Sugino Y, Misawa A, Inoue J, Kitagawa M, Hosoi H, Sugimoto T, Imoto I, Inazawa J (2007) Epigenetic silencing of prostaglandin E receptor 2 (PTGER2) is associated with progression of neuroblastomas. Oncogene. https://doi.org/10.1038/sj.onc.1210550
Taskesen E, Babaei S, Reinders MM, de Ridder J (2015) Integration of gene expression and DNA-methylation profiles improves molecular subtype classification in acute myeloid leukemia. BMC Bioinform 16(Suppl 4):S5
The Cancer Genome Atlas Research Network (2016) Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. https://doi.org/10.1056/nejmoa1505917
Thomas J, Sael L (2017) Multi-kernel LS-SVM based integration bio-clinical data analysis and application to ovarian cancer. Int J Data Mining Bioinform. https://doi.org/10.1504/ijdmb.2017.089281
Tibshirani R, Hastie T, Narasimhan B, Chu G (2002) Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.082099299
Vincent A, Omura N, Hong SM, Jaffe A, Eshleman J, Goggins M (2011) Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res 17(13):4341–4354
Wang Y, Zhao M, Liu J, Sun Z, Ni J, Liu H (2017) miRNA-125b regulates apoptosis of human non-small cell lung cancer via the PI3K/Akt/GSK3β signaling pathway. Oncol Rep 38(3):1715–1723
Wang Z, Teng D, Li Y, Hu Z, Liu L, Zheng H (2018) A six-gene-based prognostic signature for hepatocellular carcinoma overall survival prediction. Life Sci 203:83–91
Wei JH, Haddad A, Wu KJ, Zhao HW, Kapur P, Zhang ZL, Zhao LY et al (2015) A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun 6:8699
Weiskopf K (2017) Cancer immunotherapy targeting the CD47/SIRPα axis. Eur J Cancer 76:100–109
Wils LJ, Bijlsma MF (2018) Epigenetic regulation of the hedgehog and Wnt pathways in cancer. Crit Rev Oncol Hematol 121:23–44
Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, Byrnes G, Durand G, Carreira C, Michelon J et al (2013) Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS ONE 8(3):e57886
Wu C, Zhou F, Ren J, Li X, Jiang Y, Ma S (2019) A selective review of multi-level omics data integration using variable selection. High-Throughput. https://doi.org/10.3390/ht8010004
Xu Z, Jin R, Yang H, King I, Lyu MR (2010) Simple and efficient multiple kernel learning by group lasso. In: Proceedings of the 27th international conference on machine learning (ICML-10), pp 1175–1182. Citeseer
Yan JW, Lin JS, He XX (2014) The emerging role of miR-375 in cancer. Int J Cancer 135(5):1011–1018
Yan KK, Zhao H, Pang H (2017) A comparison of graph- and kernel-based –omics data integration algorithms for classifying complex traits. BMC Bioinform. https://doi.org/10.1186/s12859-017-1982-4
Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G (2014) Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26(4):577–590
Zhu B, Song N, Shen R, Arora A, Machiela MJ, Song L, Landi MT et al (2017) Integrating clinical and multiple omics data for prognostic assessment across human cancers. Sci Rep. https://doi.org/10.1038/s41598-017-17031-8
Funding
P.K.V acknowledges financial support from the Early Career Research Award Scheme, Science and Engineering Research Board, DST, India (ECR/2016/000488).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, N.P., Vinod, P.K. Integrative analysis of DNA methylation and gene expression in papillary renal cell carcinoma. Mol Genet Genomics 295, 807–824 (2020). https://doi.org/10.1007/s00438-020-01664-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01664-y