In honor of my father Leonardo Andrade Capetillo.
Abstract
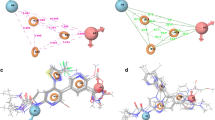
The DNA topoisomerase enzymes, Topo I and Topo II, have been used as molecular targets for drug design of anticancer agents. The search is for new anticancer therapies that respond to the toxicity of current drug treatments and tumor resistance. The present study provides insights into a likely dual inhibitory effect on Topo I and II of trifluoromethylsulfonamide (triflamide) derivatives by computational docking studies. The physicochemical properties of these compounds were evaluated by Lipinski’s rules. Molecular docking simulations were conducted to determine the possible molecular target, mode and energy binding of the triflamide derivatives. An in silico analysis indicated that the triflamide derivatives interact with amino acid residues at the active site of Topo I and Topo II. The highest binding energy for the Topo I complex was shown by 1g and for the Topo II complex by 1e; these studies were validated by the analysis of decoys. Virtual mutations of Topo I and Topo II were tested, revealing the importance of certain active site residues on the binding mode and binding energy of the test triflamide derivatives. Overall, the results suggest that the compounds 1g and 1e could be drugs promising for the future design and development of anticancer agents.
Graphic abstract
Analysis of physicochemical, toxicological and pharmacokinetic properties of triflamide derivatives and their interactions with DNA topoisomerase I and II enzymes (Topo I and II) have been reported in this study. These computational results indicated that triflamide derivatives could present a dual inhibition by binding to the active site of both enzymes, validated by decoys analysis. On the other hand, the generation of virtual mutants of Topo I and II demonstrated the great influence of certain amino acids on the mode of union of triflamides.














Similar content being viewed by others
References
Shewach D S and Kuchta R D 2009 Introduction to cancer chemotherapeutics Chem. Rev. 109 2859
Torre L A, Bray F, Siegel R L, Ferlay J, Lortet-Tieulent J and Jemal A 2015 Global Cancer Statistics 2012 CA: Cancer. J. Clin. 65 87
Ekwueme D U, Yabroff K R, Guy G P, Banegas M P, de Moor J S, Li C, Han X, Zheng Z, Soni A, Davidoff A, Rechis R and Virgo K S 2014 Medical costs and productivity losses of cancer survivors - United States, 2008-2011 Morb. Mortal. Wkly. Rep. 63 505
Badal S and Delgoda R (Eds.) 2016 Pharmacognosy: Fundamentals, Applications and Strategies (Oxford: Elsevier) p.738
Reddy-Holdcraft S, Mehta P S and Agrawal A K 2014 Paclitaxel for relapsed or recurrent HIV-associated pediatric Kaposi’s sarcoma AIDS 28 800
Akazawa H 2017 Cardiotoxicity of Cancer Chemotherapy - Mechanisms and Therapeutic Approach Gan. To. Kagaku. Ryoho. 44 2058
Kelland L 2007 Broadening the clinical use of platinum drug-based chemotherapy with new analogues satraplatin and picoplatin Expert. Opin. Investig. Drugs. 16 1009
Li F, Jiang T, Li Q and Ling X 2017 Camptothecin (CPT) and its derivatives are known to target topoisomerase I (Top1) as their mechanism of action: Did we miss something in CPT analogue molecular targets for treating human disease such as cancer? Am. J. Cancer. Res. 7 2350
Pommier Y 2006 Topoisomerase I inhibitors: Camptothecins and beyond Nat. Rev. Cancer. 6 789
Kollmannsberger C, Mross K, Jakob A, Kanz L and Bokemeyer C 1999 Topotecan - A novel topoisomerase I inhibitor: Pharmacology and clinical experience Oncology 56 1
Bracher F and Tremmel T 2017 From Lead to Drug Utilizing a Mannich Reaction: The Topotecan Story Arch. Pharm. (Weinheim). 350 8
Mancini G, D’Annessa I, Coletta A, Sanna N, Chillemi G and Desideri A 2010 Structural and dynamical effects induced by the anticancer drug topotecan on the human topoisomerase I - DNA complex PLoS One 5 10
Chikamori K, Grozav A G, Kozuki T, Grabowski D, Ganapathi R and Ganapathi M K 2010 DNA topoisomerase II enzymes as molecular targets for cancer chemotherapy Curr. Cancer. Drug. Targets 10 758
Fortune J M and Osheroff N 2000 Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice Prog. Nucleic. Acid. Res. Mol. Biol. 64 221
Li F, Ling X, Harris D, Liao J, Wang Y, Westover D, Jiang G, Xu B, Boland P M and Jin C 2017 Topoisomerase I (Top1): a major target of FL118 for its antitumor efficacy or mainly involved in its side effects of hematopoietic toxicity? Am. J. Cancer. Res. 7 370
Beck W T, Danks M K 1991 Mechanisms of resistance to drugs that inhibit DNA topoisomerases Semin. Cancer. Biol. 2 235
Gómez-García O, Gómez E, Monzón-González C, Ramírez-Apan T and Álvarez-Toledano C 2017 An efficient strategy for the synthesis of 1-(trifluoromethylsulfonamido)-propan-2-yl esters and the evaluation of their cytotoxic activity Chem. Pharm. Bull. 65 248
Scozzafava A, Owa T, Mastrolorenzo A and Supuran C T 2003 Anticancer and antiviral sulfonamides Curr. Med. Chem. 10 925
Casini A, Scozzafava A, Mastrolorenzo A and Supuran LT 2002 Sulfonamides and sulfonylated derivatives as anticancer agents Curr. Cancer. Drug. Targets. 2 55
Cumaoglu A, Dayan S, Agkaya A, Ozkul Z and Ozpozan N K 2015 Synthesis and pro-apoptotic effects of new sulfonamidederivatives via activating p38/ERK phosphorylation in cancer cells J. Enzyme. Inhib. Med. Chem. 30 413
Shoaib Ahmad Shah S, Rivera G and Ashfaq M 2012 Recent Advances in Medicinal Chemistry of Sulfonamides. Rational Design as Anti-Tumoral, Anti-Bacterial and Anti-Inflammatory Agents Mini-Reviews. Med. Chem. 13 70
Pommier Y, Leo E, Zhang H and Marchand C 2010 DNA topoisomerases and their poisoning by anticancer and antibacterial drugs Chem. Biol. 17 421
Delgado J L, Hsieh C M, Chan N L and Hiasa H 2018 Topoisomerases as anticancer targets Biochem. J. 475 373
Sander T, Freyss J, Von Korff M and Rufener C 2015 DataWarrior: An open-source program for chemistry aware data visualization and analysis J. Chem. Inf. Model. 55 460
Daina A, Michielin O and Zoete V 2017 SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules Sci. Rep. 3 42717
Furnham N, Laskowski R A and Thornton J M 2013 Abstracting knowledge from the Protein Data Bank Biopolymers 99 183
Phillips J C, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel R D, Kalé L and Schulten K 2005 Scalable molecular dynamics with NAMD J. Comput. Chem. 26 1781
Humphrey W, Dalke A and Schulten K 1996 VMD: visual molecular dynamics J. Mol. Graph. 14 33
Irwin J J and Shoichet B K 2005 ZINC–a free database of commercially available compounds for virtual screening J. Chem. Inf. Model. 45 177
Pople J A, Head-Gordon M and Fox D J 1989 Gaussian-1 Theory: A General Procedure for Prediction of Molecular-Energies J. Chem. Phys. 90 5622
Morris G M, Ruth H, Lindstrom W, Sanner M F, Belew R K, Goodsell D S and Olson A J 2009 Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility J. Comput. Chem. 30 2785
Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016
Webb B and Sali A 2014 Comparative protein structure modeling using Modeller Curr. Protoc. Bioinformatics. 47 5.6.1
Mysinger M M, Carchia M, Irwin J J and Shoichet B K 2012 Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking J. Med. Chem. 55 6582
Vraka C, Nics L, Wagner K H, Hacker M, Wadsak W and Mitterhauser M 2017 LogP, a yesterday’s value? Nucl. Med. Biol. 50 1
Trapani A, Lopedota A, Denora N, Laquintana V, Franco M, Latrofa A, Trapani G and Liso G 2005 A rapid screening tool for estimating the potential of 2-hydroxypropyl-beta-cyclodextrin complexation for solubilization purposes Int. J. Pharm. 295 163
Ertl P, Rohde B and Selzer P 2000 Fast Calculation of molecular polar surface area as a sum of fragment based contributions and its application to the prediction of drug transport properties J. Med. Chem. 43 3714
Hopkins A L, Groom C R and Alex A 2004 Ligand efficiency: A useful metric for lead selection Drug. Discov. Today 9 430
Keserü G M and Makara G M 2009 The influence of lead discovery strategies on the properties of drug candidates Nat. Rev. Drug. Discov. 8 203
Lipinski C A, Lombardo F, Dominy B W and Feeney P J 2001 Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings1 Adv. Drug. Deliv. Rev. 46 3
Champoux J J 2001 DNA topoisomerases: structure, function, and mechanism Annu. Rev. Biochem. 70 369
Pan P, Li Y, Yu H, Sun H and Hou T 2013 Molecular principle of topotecan resistance by topoisomerase I mutations through molecular modeling approaches J. Chem. Inf. Model. 53 997
Tan H, Wang G, Li J, Meng G, Liu Z, Dong M, Li Y, Ju D and Zhang Q 2015 Synthesis of novel 10-hydroxycamptothecin derivatives utilizing topotecan hydrochloride as ortho-quinonemethide precursor Bioorg. Med. Chem. 23 118
Singh S, Das T, Awasthi M, Pandey V P, Pandey B and Dwivedi U N 2016 DNA topoisomerase-directed anticancerous alkaloids: ADMET-based screening, molecular docking, and dynamics simulation Biotechnol. Appl. Biochem. 63 125
Xin L-T, Liu L, Shao C-L, Yu R L, Chen F L, Yue S J, Wang M, Guo Z L, Fan Y C, Guan H S and Wang C Y 2017 Discovery of DNA Topoisomerase I Inhibitors with Low-Cytotoxicity Based on Virtual Screening from Natural Products Mar. Drugs. 15 217
Scotti L, Mendonça F J B, Ribeiro F F, Tavares J F, da Silva M S, Barbosa Filho J M and Scotti M T 2018 Natural product inhibitors of topoisomerases: Review and docking study Curr. Protein. Pept. Sci. 19 275
Hande K R 1998 Etoposide: four decades of development of a topoisomerase II inhibitor Eur. J. Cancer. 34 1514
Wu C C, Li T K, Farh L, Lin L Y, Lin T S, Yu Y J, Yen T J, Chiang C W and Chan N L 2011 Structural Basis of Type II Topoisomerase Inhibition by the Anticancer Drug Etoposide Science 333 459
Kumar A and Bora U 2014 Molecular docking studies of curcumin natural derivatives with DNA topoisomerase I and II-DNA complexes Interdiscip. Sci. 6 285
Oksuzoglu E, Ertan-Bolelli T, Can H, Tarhan M, Ozturk K and Yildiz I 2017 Antitumor activities on HL-60 human leukemia cell line, molecular docking, and quantum-chemical calculations of some sulfonamide-benzoxazoles Artif. Cells. Nanomed. Biotechnol. 45 1388
Guianvarćh D, Duca M, Boukarim C, Kraus-Berthier L, Léonce S, Pierré A, Pfeiffer B, Renard P, Arimondo PB, Monneret C and Dauzonne D 2004 Synthesis and biological activity of sulfonamide derivatives of epipodophyllotoxin J. Med. Chem. 47 2365
Ozawa Y, Kusano K, Owa T, Yokoi A, Asada M and Yoshimatsu K 2012 Therapeutic potential and molecular mechanism of a novel sulfonamide anticancer drug, indisulam (E7070) in combination with CPT-11 for cancer treatment Cancer. Chemother. Pharmacol. 69 1844
Lavanya R 2017 Sulphonamides: A Pharmaceutical Review Int. J. Pharma. Sci. 6 1
Shanti K D, Shanti M D and Meshram J S 2016 A convenient synthesis and molecular docking study of novel sulfonamides fused with Betti’s bases as DNA Topoisomerase II inhibitors J. Comput. Methods. Mol. Des. 6 13
Ajeet S R and Kumar A 2014 Designing of Sulfanilamide/Sulfacetamide Derivatives as Human Topoisomerase II Inhibitor: A Docking Approach Am. J. Pharmacol. Sci. 2 44
Graves A P, Brenk R and Shoichet B K 2005 Decoys for Docking J. Med. Chem. 48 3714
Schmidtke P and Barril X 2010 Understanding and Predicting Druggability. A High-Throughput Method for Detection of Drug Binding Sites J. Med. Chem. 53 5858
Sader S and Wu C 2017 Computational analysis of Amsacrine resistance in human topoisomerase II alpha mutants (R487K and E571K) using homology modeling, docking and all atom molecular dynamics simulation in explicit solvent J. Mol. Graph. Model. 72 209
Acknowledgements
The authors would like to thank the Instituto Politécnico Nacional for two grants [SIP20181529 and SIP20172002] that gave financial support to the present research. We are grateful to Bruce Allan Larsen for proofreading the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
DULCE, AP., OMAR, GG. & CECILIO, ÁT. Exploring the binding mode of triflamide derivatives at the active site of Topo I and Topo II enzymes: In silico analysis and precise molecular docking. J Chem Sci 132, 50 (2020). https://doi.org/10.1007/s12039-020-1750-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12039-020-1750-2