Abstract
Key message
Aggregation across multiple networks highlights robust co-expression interactions and improves the functional connectivity of grapevine gene co-expression networks.
Abstract
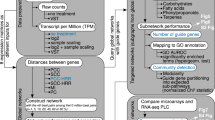
In recent years, the rapid accumulation of transcriptome datasets from diverse experimental conditions has enabled the widespread use of gene co-expression network (GCN) analysis in plants. In grapevine, GCN analysis has shown great promise for gene function prediction, however, measurable progress is currently lacking. Using accumulated microarray datasets from the grapevine whole-genome array (33 experiments, 1359 samples), we explored how meta-analysis through aggregation influences the functional connectivity (performance) of derived networks using guilt-by-association neighbor voting. Two annotation schemes, i.e. MapMan BIN and Pfam, at two sparsity thresholds, i.e. top 100 (stringent) and 300 (relaxed) ranked genes were evaluated. We observed that aggregating across multiple networks improves performance dramatically, with the aggregate outperforming the majority of functional terms across individual networks. Network sparsity and size (i.e. the number of samples and aggregates) were key factors influencing performance while the choice of annotation scheme had little. Systematic comparison with various state-of-the-art microarray and RNA-seq networks was also performed, however, none outperformed the aggregate microarray network despite having good predictive performance. Repeating these series of tests using a functional enrichment-based performance metric also showed remarkably consistent findings with guilt-by-association neighbor voting. To demonstrate its functionality, we explore the function and transcriptional regulation of grapevine EXPANSIN genes. We envisage that network aggregation will offer new and unique opportunities for gene function prediction in future grapevine functional genomics studies. To this end, we make the aggregate networks and associated metadata publicly available at VTC-Agg (https://sites.google.com/view/vtc-agg).






Similar content being viewed by others
References
Amato A, Cavallini E, Zenoni S, Finezzo L, Begheldo M, Ruperti B, Tornielli GB (2017) A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front Plant Sci 7:1979. https://doi.org/10.3389/fpls.2016.01979
Aoki Y, Okamura Y, Tadaka S, Kinoshita K, Obayashi T (2016) ATTED-II in 2016: A plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol 57:e5. https://doi.org/10.1093/pcp/pcv165
Bai M-Y, Fan M, Oh E, Wang Z-Y (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24:4917–4929. https://doi.org/10.1105/tpc.112.105163
Ballouz S, Verleyen W, Gillis J (2015) Guidance for RNA-seq co-expression network construction and analysis: Safety in numbers. Bioinformatics 31:2123–2130. https://doi.org/10.1093/bioinformatics/btv118
Ballouz S, Weber M, Pavlidis P, Gillis J (2017) EGAD: Ultra-fast functional analysis of gene networks. Bioinformatics 33:612–614. https://doi.org/10.1093/bioinformatics/btw695
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A (2013) NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 41:D991–D995. https://doi.org/10.1093/nar/gks1193
Bolger ME, Arsova B, Usadel B (2018) Plant genome and transcriptome annotations: from misconceptions to simple solutions. Brief Bioinform 19:437–449. https://doi.org/10.1093/bib/bbw135
Carvalho BS, Irizarry RA (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26:2363–2367. https://doi.org/10.1093/bioinformatics/btq431
Cosgrove DJ (2015) Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25:162–172. https://doi.org/10.1016/j.pbi.2015.05.014
Cramer GR, Ghan R, Schlauch KA, Tillett RL, Heymann H, Ferrarini A, Delledonne M, Zenoni S, Fasoli M, Pezzotti M (2014) Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol 14:1–21. https://doi.org/10.1186/s12870-014-0370-8
Dai F, Zhang C, Jiang X, Kang M, Yin X, Lu P, Zhang X, Zheng Y, Gao J (2012) RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol 160:2064–2082. https://doi.org/10.1104/pp.112.207720
Dal Santo S, Palliotti A, Zenoni S, Tornielli GB, Fasoli M, Paci P, Tombesi S, Frioni T, Silvestroni O, Bellincontro A, d’Onofrio C, Matarese F, Gatti M, Poni S, Pezzotti M (2016) Distinct transcriptome responses to water limitation in isohydric and anisohydric grapevine cultivars. BMC Genomics 17:1–19. https://doi.org/10.1186/s12864-016-3136-x
Dal Santo S, Tornielli GB, Zenoni S, Fasoli M, Farina L, Anesi A, Guzzo F, Delledonne M, Pezzotti M (2013a) The plasticity of the grapevine berry transcriptome. Genome Biol 14:r54. https://doi.org/10.1186/gb-2013-14-6-r54
Dal Santo S, Vannozzi A, Tornielli GB, Fasoli M, Venturini L, Pezzotti M, Zenoni S (2013b) Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS ONE 8:e62206. https://doi.org/10.1371/journal.pone.0062206
Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, Porceddu A, Venturini L, Bicego M, Murino V, Ferrarini A, Delledonne M, Pezzotti M (2012) The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24:3489–3505. https://doi.org/10.1105/tpc.112.100230
Gibson SM, Ficklin SP, Isaacson S, Luo F, Feltus FA, Smith MC (2013) Massive-scale gene co-expression network construction and robustness testing using random matrix theory. PLoS ONE. https://doi.org/10.1371/journal.pone.0055871
Gillis J, Pavlidis P (2012) “Guilt by association” is the exception rather than the rule in gene networks. PLoS Comput Biol 8:e1002444. https://doi.org/10.1371/journal.pcbi.1002444
Gillis J, Pavlidis P (2011) The role of indirect connections in gene networks in predicting function. Bioinformatics 27:1860–1866. https://doi.org/10.1093/bioinformatics/btr288
Giorgi FM, Del Fabbro C, Licausi F (2013) Comparative study of RNA-seq- and Microarray-derived coexpression networks in Arabidopsis thaliana. Bioinformatics 29:717–724. https://doi.org/10.1093/bioinformatics/btt053
Grimplet J, Martínez-zapater JM, Carmona MJ (2016) Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genomics 17:80. https://doi.org/10.1186/s12864-016-2398-7
Huang J, Vendramin Alegre S, Shi L, McGinnis K (2017) Construction and optimization of large gene co-expression network in maize using RNA-Seq data. Plant Physiol 175:00825.2017. https://doi.org/10.1104/pp.17.00825
Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M (2012) A Triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24:4483–4497. https://doi.org/10.1105/tpc.112.105023
Klie S, Nikoloski Z (2012) The choice between MapMan and Gene ontology for automated gene function prediction in plant science. Front Genet 3:1–14. https://doi.org/10.3389/fgene.2012.00115
Kuang JF, Chen JY, Liu XC, Han YC, Xiao YY, Shan W, Tang Y, Wu KQ, He JX, Lu WJ (2017) The transcriptional regulatory network mediated by banana (Musa acuminata) dehydration-responsive element binding (MaDREB) transcription factors in fruit ripening. New Phytol 214:762–781. https://doi.org/10.1111/nph.14389
Liesecke F, De Craene JO, Besseau S, Courdavault V, Clastre M, Vergès V, Papon N, Giglioli-Guivarc’h N, Glévarec G, Pichon O, Dugé de Bernonville T (2019) Improved gene co-expression network quality through expression dataset down-sampling and network aggregation. Sci Rep 9:1–16. https://doi.org/10.1038/s41598-019-50885-8
Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B (2014) Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ 37:1250–1258. https://doi.org/10.1111/pce.12231
Loyola R, Herrera D, Mas A, Wong DCJ, Höll J, Cavallini E, Amato A, Azuma A, Ziegler T, Aquea F, Castellarin SD, Bogs J, Tornielli GB, Peña-Neira A, Czemmel S, Alcalde JA, Matus JT, Arce-Johnson P (2016) The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J Exp Bot. https://doi.org/10.1093/jxb/erw307
Malacarne G, Coller E, Czemmel S, Vrhovsek U, Engelen K, Goremykin V, Bogs J, Moser C (2016) The grapevine VvibZIPC22 transcription factor is involved in the regulation of flavonoid biosynthesis. J Exp Bot 67:3509–3522. https://doi.org/10.1093/jxb/erw181
Malacarne G, Pilati S, Valentini S, Asnicar F, Moretto M, Sonego P, Masera L, Cavecchia V, Blanzieri E, Moser CM (2018) Discovering causal relationships in grapevine expression data to expand gene networks. A case study: four networks related to climate change. Front Plant Sci 9:1385. https://doi.org/10.3389/FPLS.2018.01385
Massonnet M, Fasoli M, Tornielli GB, Altieri M, Sandri M, Zuccolotto P, Paci P, Gardiman M, Zenoni S, Pezzotti M (2017) Ripening transcriptomic program in red and white grapevine varieties correlates with berry skin anthocyanin accumulation. Plant Physiol 174:2376–2396. https://doi.org/10.1104/pp.17.00311
Mitchell RAC, Dupree P, Shewry PR (2007) A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol 144:43–53. https://doi.org/10.1104/pp.106.094995
Moretto M, Sonego P, Pilati S, Malacarne G, Costantini L, Grzeskowiak L, Bagagli G, Grando MS, Moser C, Engelen K (2016) VESPUCCI: Exploring patterns of gene expression in grapevine. Front Plant Sci 7:633. https://doi.org/10.3389/fpls.2016.00633
Nicolas P, Lecourieux D, Gomès E, Delrot S, Lecourieux F (2013) The grape berry-specific basic helix-loop-helix transcription factor VvCEB1 affects cell size. J Exp Bot 64:991–1003. https://doi.org/10.1093/jxb/ers374
O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165:1280–1292. https://doi.org/10.1016/j.cell.2016.04.038
Obayashi T, Aoki Y, Tadaka S, Kagaya Y, Kinoshita K (2018) ATTED-II in 2018: a plant coexpression database based on investigation of the statistical property of the Mutual Rank Index. Plant Cell Physiol 59:e3. https://doi.org/10.1093/pcp/pcx191
Obayashi T, Kagaya Y, Aoki Y, Tadaka S, Kinoshita K (2019) COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res 47:D55–D62. https://doi.org/10.1093/nar/gky1155
Obayashi T, Kinoshita K (2009) Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res 16:249–260. https://doi.org/10.1093/dnares/dsp016
Ohyanagi H, Takano T, Terashima S, Kobayashi M, Kanno M, Morimoto K, Kanegae H, Sasaki Y, Saito M, Asano S, Ozaki S, Kudo T, Yokoyama K, Aya K, Suwabe K, Suzuki G, Aoki K, Kubo Y, Watanabe M, Matsuoka M, Yano K (2015) Plant omics data center: an integrated web repository for interspecies gene expression networks with NLP-based curation. Plant Cell Physiol 56:e9. https://doi.org/10.1093/pcp/pcu188
Palumbo MC, Zenoni S, Fasoli M, Massonnet M, Farina L, Castiglione F, Pezzotti M, Paci P (2014) Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell Online 26:4617–4635. https://doi.org/10.1105/tpc.114.133710
Proost S, Mutwil M (2018) CoNekT: An open-source framework for comparative genomic and transcriptomic network analyses. Nucleic Acids Res 46:W133–W140. https://doi.org/10.1093/nar/gky336
Ruprecht C, Persson S (2012) Co-expression of cell-wall related genes: new tools and insights. Front Plant Sci 3:1–7. https://doi.org/10.3389/fpls.2012.00083
Ruprecht C, Proost S, Hernandez-Coronado M, Ortiz-Ramirez C, Lang D, Rensing SA, Becker JD, Vandepoele K, Mutwil M (2017) Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J 90:447–465. https://doi.org/10.1111/tpj.13502
Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M, Kurbatova N, Malone J, Mani R, Mupo A, PedroPereira R, Pilicheva E, Rung J, Sharma A, Tang YA, Ternent T, Tikhonov A, Welter D, Williams E, Brazma A, Parkinson H, Sarkans U (2013) ArrayExpress update–trends in database growth and links to data analysis tools. Nucleic Acids Res 41:D987–D990. https://doi.org/10.1093/nar/gks1174
Savoi S, Wong DCJ, Arapitsas P, Miculan M, Bucchetti B, Peterlunger E, Fait A, Mattivi F, Castellarin SD (2016) Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.). BMC Plant Biol 16:67. https://doi.org/10.1186/s12870-016-0760-1
Savoi S, Wong DCJ, Degu A, Herrera JC, Bucchetti B, Peterlunger E, Fait A, Mattivi F, Castellarin SD (2017) Multi-omics and integrated network analyses reveal new insights into the systems relationships between metabolites, structural genes, and transcriptional regulators in developing grape berries (Vitis vinifera L.) exposed to water deficit. Front Plant Sci 8:1124. https://doi.org/10.3389/fpls.2017.01124
Schlosser J, Olsson N, Weis M, Reid K, Peng F, Lund S, Bowen P (2008) Cellular expansion and gene expression in the developing grape (Vitis vinifera L.). Protoplasma 232:255–265. https://doi.org/10.1007/s00709-008-0280-9
Serin EAR, Nijveen H, Hilhorst HWM, Ligterink W (2016) Learning from co-expression networks: possibilities and challenges. Front Plant Sci 7:1–18. https://doi.org/10.3389/fpls.2016.00444
Sun X, Matus JT, Wong DCJ, Wang Z, Chai F, Zhang L, Fang T, Zhao L, Wang Y, Han Y, Wang Q, Li S, Liang Z, Xin H (2018) The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J Exp Bot 69:1749–1764. https://doi.org/10.1093/jxb/ery020
Sun X, Zhang L, Wong DCJ, Wang Y, Zhu Z, Xu G, Wang Q, Li S, Liang Z, Xin H (2019) The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. https://doi.org/10.1111/tpj.14378
Sundell D, Street NR, Kumar M, Mellerowicz EJ, Kucukoglu M, Johnsson C, Kumar V, Mannapperuma C, Delhomme N, Nilsson O, Tuominen H, Pesquet E, Fischer U, Niittylä T, Sundberg B, Hvidsten TR (2017) AspWood: high-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29:1585–1604. https://doi.org/10.1105/tpc.17.00153
Suzuki H, Oshita E, Fujimori N, Nakajima Y, Kawagoe Y, Suzuki S (2014) Grape expansins, VvEXPA14 and VvEXPA18 promote cell expansion in transgenic Arabidopsis plant. Plant Cell Tissue Organ Cult 120:1077–1085. https://doi.org/10.1007/s11240-014-0662-6
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939. https://doi.org/10.1111/j.1365-313X.2004.02016.x
Ulitsky I, Shamir R (2009) Identifying functional modules using expression profiles and confidence-scored protein interactions. Bioinformatics 25:1158–1164. https://doi.org/10.1093/bioinformatics/btp118
Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ (2009) Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32:1633–1651. https://doi.org/10.1111/j.1365-3040.2009.02040.x
Vannozzi A, Wong DCJ, Holl J, Hmmam I, Matus JT, Bogs J, Ziegler T, Dry I, Barcaccia G, Lucchin M (2018) Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol 59:1043–1059. https://doi.org/10.1093/pcp/pcy045
Verleyen W, Ballouz S, Gillis J (2015) Measuring the wisdom of the crowds in network-based gene function inference. Bioinformatics 31:745–752. https://doi.org/10.1093/bioinformatics/btu715
Vitulo N, Forcato C, Carpinelli EC, Telatin A, Campagna D, D’Angelo M, Zimbello R, Corso M, Vannozzi A, Bonghi C, Lucchin M, Valle G (2014) A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol 14:99. https://doi.org/10.1186/1471-2229-14-99
Wong DCJ, Lopez Gutierrez R, Gambetta GA, Castellarin SD (2017) Genome-wide analysis of cis-regulatory element structure and discovery of motif-driven gene co-expression networks in grapevine. DNA Res 24:311–326. https://doi.org/10.1093/dnares/dsw061
Wong DCJ, Matus JT (2017) Constructing integrated networks for identifying new secondary metabolic pathway regulators in grapevine : Recent applications and future opportunities. Front Plant Sci 8:505. https://doi.org/10.3389/fpls.2017.00505
Wong DCJ, Schlechter R, Vannozzi A, Höll J, Hmmam I, Bogs J, Tornielli GB, Castellarin SD, Matus JT (2016) A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res 23:451–466. https://doi.org/10.1093/dnares/dsw028
Wong DCJ, Sweetman C, Drew DP, Ford CM (2013) VTCdb: a gene co-expression database for the crop species Vitis vinifera (grapevine). BMC Genomics 14:882. https://doi.org/10.1186/1471-2164-14-882
Wong DCJ, Zhang L, Merlin I, Castellarin SD, Gambetta GA (2018) Structure and transcriptional regulation of the major intrinsic protein gene family in grapevine. BMC Genomics 19:248. https://doi.org/10.1186/s12864-018-4638-5
Zenoni S, Fasoli M, Guzzo F, Dal Santo S, Amato A, Anesi A, Commisso M, Herderich M, Ceoldo S, Avesani L, Pezzotti M, Tornielli GB (2016) Disclosing the molecular basis of the postharvest life of berry in different grapevine genotypes. Plant Physiol 172:1821–1843. https://doi.org/10.1104/pp.16.00865
Acknowledgements
The author would like to thank the anonymous reviewers for their helpful and constructive comments, José Tomás Matus for critically reviewing this work in its early stages, Marek Mutwil for the provision of the grape CoNekT dataset, and the grapevine research community for making various RNA-seq and microarray datasets publicly available.
Author information
Authors and Affiliations
Contributions
DCJW conceived the study, compiled and analysed the microarray data, performed the analysis, and drafted the manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The author declares that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wong, D.C.J. Network aggregation improves gene function prediction of grapevine gene co-expression networks. Plant Mol Biol 103, 425–441 (2020). https://doi.org/10.1007/s11103-020-01001-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01001-2