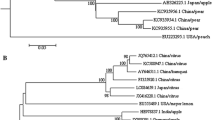
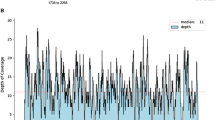
Abstract
We provide the complete sequence of a virus tentatively named “Tetranychus urticae-associated picorna-like virus 1PK13” (TuaPV1-PK13) obtained from the high-throughput sequencing of a symptomless apple leaf sample. Although the virus sequence was originally derived from apple leaves, the data suggest that the virus is associated with the two-spotted mite Tetranychus urticae.


Similar content being viewed by others
References
Simmonds P, Adams MJ, Benkő M et al (2017) Consensus statement: Virus taxonomy in the age of metagenomics. Nat Rev Micro 15:161–168. https://doi.org/10.1038/nrmicro.2016.177
Desselberger U (2018) Virus taxonomy—a taxing task. Arch Virol 163:2019–2020. https://doi.org/10.1007/s00705-018-3933-4
François S, Mutuel D, Duncan A et al (2019) A new prevalent densovirus discovered in Acari. Insight from metagenomics in viral communities associated with two-spotted mite (Tetranychus urticae) populations. Viruses 11:E233. https://doi.org/10.3390/v11030233
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Camacho C, Coulouris G, Avagyan V et al (2009) BLAST+: architecture and applications. BMC Bioinform 10:421. https://doi.org/10.1186/1471-2105-10-421
Milusheva S, Phelan J, Piperkova N et al (2018) Molecular analysis of the complete genome of an unusual virus detected in sweet cherry (Prunus avium) in Bulgaria. Eur J Plant Pathol 153:197–207. https://doi.org/10.1007/s10658-018-1555-z
Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE (2008) Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch Virol 153:715–727. https://doi.org/10.1007/s00705-008-0041-x
Migeon A, Nouguier E, Dorkeld F (2010) Spider mites web: a comprehensive database for the Tetranychidae. Trends in Acarology, 4th edn. Springer, Dordrecht, pp 557–560
Acknowledgements
We kindly thank Alena Matyášová for her technical assistance, and Ing. Jana Suchá, Research and Breeding Institute of Pomology (Holovousy, Czech Republic), for the PK13 apple sample. This study was funded by the project QK1910065 of the Czech Ministry of Agriculture and by the institutional support RVO60077344.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Handling Editor: Tim Skern.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koloniuk, I., Přibylová, J. & Špak, J. Complete genome sequence of a mite-associated virus obtained by high-throughput sequencing analysis of an apple leaf sample. Arch Virol 165, 1501–1504 (2020). https://doi.org/10.1007/s00705-020-04620-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04620-8