Abstract
The endosomal sorting complexes required for transport (ESCRTs) mediate diverse membrane remodeling events. These typically require ESCRT-III proteins to stabilize negatively curved membranes; however, recent work has indicated that certain ESCRT-IIIs also participate in positive-curvature membrane-shaping reactions. ESCRT-IIIs polymerize into membrane-binding filaments, but the structural basis for negative versus positive membrane remodeling by these proteins remains poorly understood. To learn how certain ESCRT-IIIs shape positively curved membranes, we determined structures of human membrane-bound CHMP1B-only, membrane-bound CHMP1B + IST1, and IST1-only filaments by cryo-EM. Our structures show how CHMP1B first polymerizes into a single-stranded helical filament, shaping membranes into moderate-curvature tubules. Subsequently, IST1 assembles a second strand on CHMP1B, further constricting the membrane tube and reducing its diameter nearly to the fission point. Each step of constriction thins the underlying bilayer, lowering the barrier to membrane fission. Our structures reveal how a two-component, sequential polymerization mechanism drives membrane tubulation, constriction and bilayer thinning.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Cryo-EM maps and models were deposited to the PDB and EMDB with the following codes: membrane-bound CHMP1B-only filament (PDB 6TZ9, EMD-20590), membrane-bound right-handed CHMP1B + IST1 filament (PDB 6TZ4, EMD-20588), membrane-bound left-handed CHMP1B + IST1 filament (PDB 6TZ5, EMD-20589), IST1NTDR16E K27E filament (PDB 6TZA, EMD-20591).
References
McCullough, J., Colf, L. A. & Sundquist, W. I. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82, 663–692 (2013).
Sundquist, W. I. & Krausslich, H. G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2, a006924 (2012).
Olmos, Y., Hodgson, L., Mantell, J., Verkade, P. & Carlton, J. G. ESCRT-III controls nuclear envelope reformation. Nature 522, 236–239 (2015).
Vietri, M. et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231–235 (2015).
Raab, M. et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 (2016).
Denais, C. M. et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016).
Olmos, Y., Perdrix-Rosell, A. & Carlton, J. G. Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol. 26, 2635–2641 (2016).
Skowyra, M. L., Schlesinger, P. H., Naismith, T. V. & Hanson, P. I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 (2018).
Chang, C. L. et al. Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J. Cell Biol. 218, 2583–2599 (2019).
Mast, F. D. et al. ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol. 217, 2087–2102 (2018).
McCullough, J., Frost, A. & Sundquist, W. I. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu. Rev. Cell Dev. Biol. 34, 85–109 (2018).
Allison, R. et al. Defects in ER-endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 216, 1337–1355 (2017).
McCullough, J. et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548–1551 (2015).
Allison, R. et al. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol. 202, 527–543 (2013).
Bajorek, M. et al. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 16, 754–762 (2009).
Muziol, T. et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10, 821–830 (2006).
Xiao, J. et al. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell 20, 3514–3524 (2009).
Hanson, P. I., Roth, R., Lin, Y. & Heuser, J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389–402 (2008).
Lata, S. et al. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 378, 818–827 (2008).
Lin, Y., Kimpler, L. A., Naismith, T. V., Lauer, J. M. & Hanson, P. I. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 280, 12799–12809 (2005).
Tang, S. et al. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. Elife 4, e12548 (2015).
McMillan, B. J. et al. Electrostatic interactions between elongated monomers drive filamentation of Drosophila Shrub, a metazoan ESCRT-III protein. Cell Rep. 16, 1211–1217 (2016).
Lee, I. H., Kai, H., Carlson, L. A., Groves, J. T. & Hurley, J. H. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc. Natl Acad. Sci. USA 112, 15892–15897 (2015).
Crespo-Yanez, X. et al. CHMP1B is a target of USP8/UBPY regulated by ubiquitin during endocytosis. PLoS Genet. 14, e1007456 (2018).
Stoorvogel, W., Oorschot, V. & Geuze, H. J. A novel class of clathrin-coated vesicles budding from endosomes. J. Cell Biol. 132, 21–33 (1996).
Connell, J. W. et al. ESCRT-III-associated proteins and spastin inhibit protrudin-dependent polarised membrane traffic. Cell. Mol. Life Sci. https://doi.org/10.1007/s00018-019-03313-z (2019).
Manni, M. M. et al. Acyl chain asymmetry and polyunsaturation of brain phospholipids facilitate membrane vesiculation without leakage. Elife 7, e34394 (2018).
Pinot, M. et al. Lipid cell biology. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science 345, 693–697 (2014).
Cashikar, A. G. et al. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife 3, e02184 (2014).
Wang, F. et al. A structural model of flagellar filament switching across multiple bacterial species. Nat. Commun. 8, 960 (2017).
Talledge, N. et al. The ESCRT-III proteins IST1 and CHMP1B assemble around nucleic acids. Preprint at bioRxiv https://doi.org/10.1101/386532 (2018).
Schoneberg, J., Lee, I. H., Iwasa, J. H. & Hurley, J. H. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5−17 (2017).
Chiaruttini, N. et al. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163, 866–879 (2015).
Mierzwa, B. E. et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19, 787–798 (2017).
Goliand, I. et al. Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM. Cell Rep. 24, 1756–1764 (2018).
Guizetti, J. et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620 (2011).
Banjade, S., Tang, S., Shah, Y. H. & Emr, S. D. Electrostatic lateral interactions drive ESCRT-III heteropolymer assembly. Elife 8, e46207 (2019).
Pfitzner, A.-K., Mercier, V. & Roux, A. Vps4 triggers sequential subunit exchange in ESCRT-III polymers that drives membrane constriction and fission. Preprint at bioRxiv https://doi.org/10.1101/718080 (2019).
Rajamoorthi, K., Petrache, H. I., McIntosh, T. J. & Brown, M. F. Packing and viscoelasticity of polyunsaturated ω-3 and ω-6 lipid bilayers as seen by 2H NMR and X-ray diffraction. J. Am. Chem. Soc. 127, 1576–1588 (2005).
Lipowsky, R. Coupling of bending and stretching deformations in vesicle membranes. Adv. Colloid Interface Sci. 208, 14–24 (2014).
Rozycki, B. & Lipowsky, R. Spontaneous curvature of bilayer membranes from molecular simulations: asymmetric lipid densities and asymmetric adsorption. J. Chem. Phys. 142, 054101 (2015).
Allain, J. M., Storm, C., Roux, A., Ben Amar, M. & Joanny, J. F. Fission of a multiphase membrane tube. Phys. Rev. Lett. 93, 158104 (2004).
Liu, J., Kaksonen, M., Drubin, D. G. & Oster, G. Endocytic vesicle scission by lipid phase boundary forces. Proc. Natl Acad. Sci. USA 103, 10277–10282 (2006).
Chiaruttini, N. & Roux, A. Dynamic and elastic shape transitions in curved ESCRT-III filaments. Curr. Opin. Cell Biol. 47, 126–135 (2017).
Lenz, M., Morlot, S. & Roux, A. Mechanical requirements for membrane fission: common facts from various examples. FEBS Lett. 583, 3839–3846 (2009).
Fabrikant, G. et al. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 5, e1000575 (2009).
Maity, S. et al. VPS4 triggers constriction and cleavage of ESCRT-III helical filaments. Sci. Adv. 5, eaau7198 (2019).
Henne, W. M., Buchkovich, N. J. & Emr, S. D. The ESCRT pathway. Dev. Cell 21, 77–91 (2011).
Peel, S., Macheboeuf, P., Martinelli, N. & Weissenhorn, W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem. Sci. 36, 199–210 (2011).
von Filseck, J. M. et al. Anisotropic ESCRT-III architecture governs helical membrane tube formation. Preprint at bioRxiv https://doi.org/10.1101/716308 (2019).
Harker-Kirschneck, L., Baum, B. & Saric, A. E. Changes in ESCRT-III filament geometry drive membrane remodelling and fission in silico. BMC Biol. 17, 82 (2019).
Schoneberg, J. et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 362, 1423–1428 (2018).
Samson, R. Y., Obita, T., Freund, S. M., Williams, R. L. & Bell, S. D. A role for the ESCRT system in cell division in archaea. Science 322, 1710–1713 (2008).
Stuchell-Brereton, M. D. et al. ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744 (2007).
Yang, D. et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15, 1278–1286 (2008).
Chernomordik, L. V. & Kozlov, M. M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 15, 675–683 (2008).
Lai, A. L., Park, H., White, J. M. & Tamm, L. K. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J. Biol. Chem. 281, 5760–5770 (2006).
Bertin, A. et al. Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Preprint at bioRxiv https://doi.org/10.1101/847319 (2019).
Agromayor, M. et al. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell 20, 1374–1387 (2009).
Bajorek, M. et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 20, 1360–1373 (2009).
Andersen, K. R., Leksa, N. C. & Schwartz, T. U. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins 81, 1857–1861 (2013).
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Morin, A. et al. Collaboration gets the most out of software. Elife 2, e01456 (2013).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
He, S. & Scheres, S. H. W. Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017).
Desfosses, A., Ciuffa, R., Gutsche, I. & Sachse, C. SPRING - an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J. Struct. Biol. 185, 15–26 (2014).
Egelman, E. H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Bio.l 74, 531–544 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Bergman, J., Osunbayo, O. & Vershinin, M. Constructing 3D microtubule networks using holographic optical trapping. Sci. Rep. 5, 18085 (2015).
Romanov, V., McCullough, J., Gale, B. K. & Frost, A. A Tunable microfluidic device enables cargo encapsulation by cell- or organelle-sized lipid vesicles comprising asymmetric lipid bilayers. Adv. Biosyst. 3, 1900010 (2019).
Smolsky, I. L. et al. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J. Appl. Crystallogr. 40, S453–S458 (2007).
Brzustowicz, M. R. & Brunger, A. T. X-ray scattering from unilamellar lipid vesicles. J. Appl. Crystallogr. 38, 126–131 (2005).
Moss, F. R. III et al. Ladderane phospholipids form a densely packed membrane with normal hydrazine and anomalously low proton/hydroxide permeability. Proc. Natl Acad. Sci. USA 115, 9098–9103 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank: members of the Frost lab for helpful discussion, especially P. Thomas and M. Sun with computational assistance; M. Grabe, M. Tucker, D. Argudo, E. Lyman, A. Sodt, G. Huber and A. Roux for discussions on membrane biophysical behavior; T. Weiss for assistance with SAXS data collection; M. Braunfeld, D. Bulkley, M. Harrington, A. Myasnikov and Z. Yu of the UCSF Center for Advanced CryoEM for microscopy support; and J. Baker-LePain and the QB3 shared cluster (NIH grant 1S10OD021596-01) for computational support. Structural biology applications used in this project were compiled and configured by SBGrid. The Titan X Pascal used for this research was donated by the NVIDIA Corporation. This work was supported by NSF grant ENG-1563280 (to M.D.V.) and NIH grants P50 AI150464 and 1DP2GM110772-01 (to A.F.), R01 GM112080 and R37 AI51174 (to W.I.S.). A.F. is also supported by a Faculty Scholar grant from the HHMI and is a Chan Zuckerberg Biohub investigator.
Author information
Authors and Affiliations
Contributions
H.C.N. performed biochemical and cryo-EM experiments and analysis. N.T. assisted with cryo-EM experiments and analysis. J.M. contributed to experiments and discussions. A.S. performed optical trap experiments and analysis. F.R.M. performed SAXS experiments and analysis. J.H.I. contributed illustrations and animations as well as discussions. M.D.V., W.I.S. and A.F. supervised all work. H.C.N., W.I.S. and A.F. prepared the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CryoEM validation of membrane-bound CHMP1B-only filament.
a, Angular distribution of the membrane-bound CHMP1B filament. b, Half map Fourier shell correlation (FSC) of the membrane-bound CHMP1B filament. c, Map to model FSC of the membrane-bound CHMP1B filament.
Extended Data Fig. 2 CryoEM reconstruction of the membrane-bound CHMP1B + IST1 filament at higher curvature and comparison of left- and right-handed CHMP1B + IST1 filaments.
a, CryoEM 3D reconstruction of the membrane-bound left-handed CHMP1B + IST1 filament. End-on view down the helical axis in grey-scale (left) or colored (middle). Right, internal view looking outward from the membrane surface along the helical axis. IST1 protomers (cyan) bind to the exterior of CHMP1B (green), leading to constriction of the membrane (grey). IST1 and CHMP1B protomers are highlighted in dark cyan and green, respectively. Diameters of the entire tube and membrane leaflet peak-to-peak distances are annotated. b, Electron density maps of CHMP1B from the left-handed (left) or right-handed (right) membrane-bound CHMP1B + IST1 filaments. Five copies of CHMP1B are shown as ribbons. c, Superposition of a CHMP1B protomer from the left-handed (purple) and right-handed (green) CHMP1B + IST1 filaments aligned to the CHMP1B N-terminal α1-α2 helices (left) or C-terminal α4-α5 helices (right).
Extended Data Fig. 3 Local resolution estimates and cryoEM validation of membrane-bound CHMP1B + IST1 filaments.
a, Angular distribution of right-handed (top) and left-handed (bottom) membrane-bound CHMP1B + IST1 filaments. b, Half map FSCs of right-handed (top) and left-handed (bottom) CHMP1B + IST1 filaments. c, Map to model FSCs right-handed (top) and left-handed (bottom) CHMP1B + IST1 filaments. d, Local resolution estimates of right-handed (left) and left-handed (right) CHMP1B + IST1 filaments.
Extended Data Fig. 4 IST1 does not discriminate between left- or right-handed CHMP1B filaments.
a, Electron density maps of the IST1 strands from the right-handed (left) or left-handed (right) membrane-bound CHMP1B + IST1 filaments. Two subunits of IST1 are shown as ribbons. b, Superposition of CHMP1B and IST1 protomers from the right-handed (dark green and dark blue, respectively) and left-handed (light green and cyan, respectively) CHMP1B + IST1 filaments. c, Superposition as in (b) but only with the C-terminal region of CHMP1B and IST1. d, Superposition of two subunits of IST1 from the right- or left-handed copolymers from (a).
Extended Data Fig. 5 Real-time monitoring of CHMP1B and IST1 membrane constriction and elongation.
a-c, Still images representing deformation of two membrane tubes due to transverse flow of (a) buffer alone, (b) then 0.5 μM CHMP1B, (c) and a final addition of 0.5 μM IST1. Solid arrows in (a-c) highlight tubule locations. d-e, Contours of lower (d) and upper (e) membrane tubes extracted from panels (a-c) showing the extension of the tubes upon addition of CHMP1B and IST1.
Extended Data Fig. 6 CryoEM validation of the IST1NTDR16E K27E filament.
a, Angular distribution of the IST1NTDR16E K27E filament. b, Half map FSC of the IST1NTDR16E K27E filament. c, Map to model FSC of the IST1NTDR16E K27E filament.
Extended Data Fig. 7 Steric clashing between the CHMP1B MIM and inter-turn IST1 subunits would prevent IST1 from achieving its preferred curvature in the copolymer.
a, External view of the IST1NTDR16E K27E filament with one CHMP1B MIM (shown as a green cylinder) docked onto the IST1NTDR16E K27E j subunit. b, Zoomed in view of boxed area in (a) highlighting how the CHMP1B MIM clashes with the IST1 j + 14 subunit.
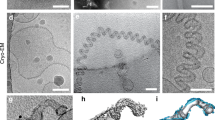
Extended Data Fig. 8 Subtle deformations in the outer leaflet observed in the moderately constricted CHMP1B-only filaments.
Left, central slice along the helical axis of the membrane-bound CHMP1B-only tubule. Right, zoomed view of boxed area in left showing very little dimpling in the outer leaflet (black dashed curved line) of the bilayer. A CHMP1B helix α1, which sits against the membrane, is highlighted in dark green.
Extended Data Fig. 9 SAXS analysis of liposomes and calculation of bilayer thickness.
a, The small angle scattering intensities for protein-free unilamellar vesicles used in this study. The black line represents the fit to the model. The blue data points were used for fitting. b, Fit results for the liposomes and the resulting thickness, D (Å). The bilayer center, ε2, was fixed at 0, and the magnitude of the central peak, ρ2, was fixed at -1. Data are mean ± s.d.
Supplementary information
Supplementary Video 1
Elbow flexing of one CHMP1B subunit.
Supplementary Video 2
Flexing of a full turn of CHMP1B subunits from low to high constriction.
Supplementary Video 3
Real-time recording of membrane tube elongation by CHMP1B and IST1.
Supplementary Video 4
IST1 alone does not promote membrane tube elongation.
Supplementary Video 5
Swinging of two IST1 subunits from initial binding to the CHMP1B filament to the constricted state (side view).
Supplementary Video 6
Swinging of two IST1 subunits from initial binding to the CHMP1B filament to the constricted state (top-down view). CHMP1B and the membrane would lie at the top of the animation.
Supplementary Video 7
Constriction of a membrane tubule by CHMP1B and IST1.
Rights and permissions
About this article
Cite this article
Nguyen, H.C., Talledge, N., McCullough, J. et al. Membrane constriction and thinning by sequential ESCRT-III polymerization. Nat Struct Mol Biol 27, 392–399 (2020). https://doi.org/10.1038/s41594-020-0404-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0404-x
This article is cited by
-
Das ESCRT-III-Homolog VIPP1 vermittelt Thylakoid-Biogenese und -Erhaltung
BIOspektrum (2024)
-
An ESCRT grommet cooperates with a diffusion barrier to maintain nuclear integrity
Nature Cell Biology (2023)
-
Structural basis of CHMP2A–CHMP3 ESCRT-III polymer assembly and membrane cleavage
Nature Structural & Molecular Biology (2023)
-
Brominated lipid probes expose structural asymmetries in constricted membranes
Nature Structural & Molecular Biology (2023)
-
Cryo-electron tomography reveals structural insights into the membrane remodeling mode of dynamin-like EHD filaments
Nature Communications (2022)