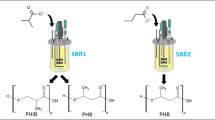
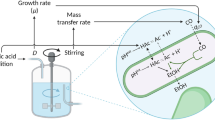
Abstract
Isobutyrate (i-butyrate) is a versatile platform chemical, whose acid form is used as a precursor of plastic and emulsifier. It can be produced microbially either using genetically engineered organisms or via microbiomes, in the latter case starting from methanol and short-chain carboxylates. This opens the opportunity to produce i-butyrate from non-sterile feedstocks. Little is known on the ecology and process conditions leading to i-butyrate production. In this study, we steered i-butyrate production in a bioreactor fed with methanol and acetate under various conditions, achieving maximum i-butyrate productivity of 5.0 mM day−1, with a concurrent production of n-butyrate of 7.9 mM day−1. The production of i-butyrate was reversibly inhibited by methanogenic inhibitor 2-bromoethanesulfonate. The microbial community data revealed the co-dominance of two major OTUs during co-production of i-butyrate and n-butyrate in two distinctive phases throughout a period of 54 days and 28 days, respectively. The cross-comparison of product profile with microbial community composition suggests that the relative abundance of Clostridium sp. over Eubacterium sp. is correlated with i-butyrate productivity over n-butyrate productivity.






Similar content being viewed by others
Data availability
All relevant raw fastq files that were used for the microbial community analysis in this study have been deposited in the National Center for Biotechnology Information (NCBI) database (BioProject accession number PRJNA572585).
References
Acosta N, De Vrieze J, Sandoval V, Sinche D, Wierinck I, Rabaey K (2018) Cocoa residues as viable biomass for renewable energy production through anaerobic digestion. Bioresour Technol 265:568–572. https://doi.org/10.1016/j.biortech.2018.05.100
Aguilar A, Casas C, Lafuente J, Lema JM (1990) Kinetic modelling of isomerization and anaerobic degradation of n- and i-butyrate. J Ferment Bioeng 69(4):261–264. https://doi.org/10.1016/0922-338X(90)90225-L
Allison MJ (1978) Production of branched-chain volatile fatty acids by certain anaerobic bacteria. Appl Environ Microbiol 35(5):872–877
Allison MJ, Bryant MP (1963) Biosynthesis of branched-chain amino acids from branched-chain fatty acids by rumen bacteria. Arch Biochem Biophys 101(2):269–277. https://doi.org/10.1016/S0003-9861(63)80012-0
Andersen SJ, Hennebel T, Gildemyn S, Coma M, Desloover J, Berton J, Tsukamoto J, Stevens C, Rabaey K (2014) Electrolytic membrane extraction enables production of fine chemicals from biorefinery sidestreams. Environ Sci Technol 48(12):7135–7142. https://doi.org/10.1021/es500483w
Andersen SJ, De Groof V, Khor WC, Roume H, Props R, Coma M, Rabaey K (2017) A Clostridium group iV species dominates and suppresses a mixed culture fermentation by tolerance to medium chain fatty acids products. Front Bioeng Biotechnol 5:8. https://doi.org/10.3389/fbioe.2017.00008
Angelidaki I, Ahring BK (1995) Isomerization of n- and i-butyrate in anaerobic methanogenic systems. Antonie Van Leeuwenhoek 68(4):285–291. https://doi.org/10.1007/BF00874138
Balan V, Chiaramonti D, Kumar S (2013) Review of US and EU initiatives toward development, demonstration, and commercialization of lignocellulosic biofuels. Biofuels Bioprod Biorefin 7(6):732–759. https://doi.org/10.1002/bbb.1436
Behrens M, Studt F, Kasatkin I, Kühl S, Hävecker M, Abild-Pedersen F, Zander S, Girgsdies F, Kurr P, Kniep B-L (2012) The active site of methanol synthesis over Cu/ZnO/Al2O3 industrial catalysts. Science 336(6083):893–897
Boone DR (2015) Methanobacteria class. nov. Bergey’s Manual of systematics of archaea and bacteria:1–1. https://doi.org/10.1002/9781118960608.cbm00027
Brendelberger G, Rétey J, Ashworth DM, Reynolds K, Willenbrock F, Robinson JA (1988) The enzymic interconversion of isobutyryl and n-butyrylcarba (dethia)-coenzyme a: a coenzyme-B12-dependent carbon skeleton rearrangement. Angew Chem Int Ed Eng 27(8):1089–1090. https://doi.org/10.1002/anie.198810891
Candry P, Van Daele T, Denis K, Amerlinck Y, Andersen SJ, Ganigué R, Arends JB, Nopens I, Rabaey K (2018) A novel high-throughput method for kinetic characterisation of anaerobic bioproduction strains, applied to Clostridium kluyveri. Sci Rep 8(1):9724. https://doi.org/10.1038/s41598-018-27594-9
Carvajal-Arroyo JM, Candry P, Andersen SJ, Props R, Seviour TW, Ganigué R, Rabaey K (2019) Granular fermentation enables high rate caproic acid production from solid-free thin stillage. Green Chem 21(6):1330–1339. https://doi.org/10.1039/c8gc03648a
Chen S, Dong X (2005) Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int J Syst Evol Microbiol 55(6):2257–2261. https://doi.org/10.1099/ijs.0.63807-0
Chen W, Ye Y, Steinbusch K, Strik D, Buisman C (2016a) Methanol as an alternative electron donor in chain elongation for butyrate and caproate formation. Biomass Bioenergy 93:201–208. https://doi.org/10.1016/j.biombioe.2016.07.008
Chen WS, Huang S, Strik DP, Buisman CJ (2016b) Isobutyrate biosynthesis via methanol chain elongation: converting organic wastes to platform chemicals. J Chem Technol Biotechnol 92(6):1370–1379. https://doi.org/10.1002/jctb.5132
Chen W-S, Strik DP, Buisman CJ, Kroeze C (2017) Production of caproic acid from mixed organic waste-an environmental life cycle perspective. Environ Sci Technol 51(12):7159–7168. https://doi.org/10.1021/acs.est.6b06220
Coma M, Vilchez-Vargas R, Roume H, Jauregui R, Pieper DH, Rabaey K (2016) Product diversity linked to substrate usage in chain elongation by mixed-culture fermentation. Environ Sci Technol 50(12):6467–6476. https://doi.org/10.1021/acs.est.5b06021
Daniell J, Köpke M, Simpson S (2012) Commercial biomass syngas fermentation. Energies 5(12):5372–5417. https://doi.org/10.3390/en5125372
de Smit SM, de Leeuw KD, Buisman CJN, Strik DPBTB (2019) Continuous n-valerate formation from propionate and methanol in an anaerobic chain elongation open-culture bioreactor. Biotechnol Biofuels 12(1):132. https://doi.org/10.1186/s13068-019-1468-x
Demirbas A (2008) Biomethanol production from organic waste materials. Energ Source Part A 30(6):565–572. https://doi.org/10.1080/15567030600817167
Florencio L, Field JA, Lettinga G (1995) Substrate competition between methanogens and acetogens during the degradation of methanol in UASB reactors. Water Res 29(3):915–922. https://doi.org/10.1016/0043-1354(94)00199-H
Gantner S, Andersson AF, Alonso-Sáez L, Bertilsson S (2011) Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J Microbiol Methods 84(1):12–18. https://doi.org/10.1016/j.mimet.2010.10.001
Genthner BS, Bryant M (1982) Growth of Eubacterium limosum with carbon monoxide as the energy source. Appl Environ Microbiol 43(1):70–74
Genthner S, Bryant M (1987) Additional characteristics of one-carbon-compound utilization by Eubacterium limosum and Acetobacterium woodii. Appl Environ Microbiol 53(3):471–476
Genthner B, Davis C, Bryant MP (1981) Features of rumen and sewage sludge strains of Eubacterium limosum, a methanol-and H2-CO2-utilizing species. Appl Environ Microbiol 42(1):12–19
Gunsalus RP, Romesser JA, Wolfe RS (1978) Preparation of coenzyme M analogs and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry 17(12):2374–2377. https://doi.org/10.1021/bi00605a019
Hugenholtz J, Ljungdahl LG (1990) Metabolism and energy generation in homoacetogenic Clostridia. FEMS Microbiol Rev 7(3–4):383–389. https://doi.org/10.1111/j.1574-6968.1990.tb04941.x
Ingólfsson HI, Andersen OS (2011) Alcohol’s effects on lipid bilayer properties. Biophys J 101(4):847–855. https://doi.org/10.1016/j.bpj.2011.07.013
Jarboe LR, Royce LA, Liu P (2013) Understanding biocatalyst inhibition by carboxylic acids. Front Microbiol 4:272. https://doi.org/10.3389/fmicb.2013.00272
Jeong J, Bertsch J, Hess V, Choi S, Choi I-G, Chang IS, Müller V (2015) Energy conservation model based on genomic and experimental analyses of a carbon monoxide-utilizing, butyrate-forming acetogen, Eubacterium limosum KIST612. Appl Environ Microbiol 81(14):4782–4790. https://doi.org/10.1128/AEM.00675-15
Junicke H, Van Loosdrecht M, Kleerebezem R (2016) Kinetic and thermodynamic control of butyrate conversion in non-defined methanogenic communities. Appl Microbiol Biotechnol 100(2):915–925. https://doi.org/10.1007/s00253-015-6971-9
Kleerebezem R, van Loosdrecht MC (2007) Mixed culture biotechnology for bioenergy production. Curr Opin Biotechnol 18(3):207–212. https://doi.org/10.1016/j.copbio.2007.05.001
Koytsoumpa MJD, Kakaras E, Schmidt S, Deierling A (2019) A technology review and cost analysis of the production of low carbon methanol and following methanol to gasoline process. In: W M (ed) Zukünftige Kraftstoffe. Springer Vieweg, Berlin, pp 433–463
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79(17):5112–5120. https://doi.org/10.1128/AEM.01043-13
Lang K, Zierow J, Buehler K, Schmid A (2014) Metabolic engineering of Pseudomonas sp. strain VLB120 as platform biocatalyst for the production of isobutyric acid and other secondary metabolites. Microb Cell Factories 13(1):2. https://doi.org/10.1186/1475-2859-13-2
Loubiere P, Goma G, Lindley N (1990) A non-passive mechanism of butyrate excretion operates during acidogenic fermentation of methanol by Eubacterium limosum. Antonie Van Leeuwenhoek 57(2):83–89. https://doi.org/10.1007/BF00403159
Lovley DR, Klug MJ (1982) Intermediary metabolism of organic matter in the sediments of a eutrophic lake. Appl Environ Microbiol 43(3):552–560
Madden T (2013) The BLAST sequence analysis tool. National Center for Biotechnology Information. https://www.unmc.edu/bsbc/docs/NCBI_blast.pdf. Accessed 29 Sept 2019
Martiny AC (2019) High proportions of bacteria are culturable across major biomes. ISME J (13):2125–2128. https://doi.org/10.1038/s41396-019-0410-3
Matthies C, Schink B (1992) Reciprocal isomerization of butyrate and isobutyrate by the strictly anaerobic bacterium strain WoG13 and methanogenic isobutyrate degradation by a defined triculture. Appl Environ Microbiol 58(5):1435–1439
Mountfort DO, Grant WD, Clarke R, Asher RA (1988) Eubacterium callanderi sp. nov. that demethoxylates O-methoxylated aromatic acids to volatile fatty acids. Int J Syst Evol Microbiol 38(3):254–258. https://doi.org/10.1099/00207713-38-3-254
Oksanen J (2015) Multivariate analysis of ecological communities in R: vegan tutorial. University of Oulu. http://cc.oulu.fi/~jarioksa/opetus/metodi/vegantutor.pdf. Accessed 29 Sept 2019
Oude Elferink SJ, Lens PN, Dijkema C, Stams AJ (1996) Isomerization of butyrate to isobutyrate by Desulforhabdus amnigenus. FEMS Microbiol Lett 142(2–3):237–241. https://doi.org/10.1111/j.1574-6968.1996.tb08436.x
Pacaud S, Loubiere P, Goma G (1985) Methanol metabolism by Eubacterium limosum B2: effects of pH and carbon dioxide on growth and organic acid production. Curr Microbiol 12(5):245–250. https://doi.org/10.1007/Bf01567972
Pacaud S, Loubiere P, Goma G, Lindley N (1986a) Effects of various organic acid supplements on growth rates of Eubacterium limosum B2 on methanol. Appl Microbiol Biotechnol 24(1):75–78
Pacaud S, Loubiere P, Goma G, Lindley N (1986b) Organic acid production during methylotrophic growth of Eubacterium limosum B2: displacement towards increased butyric acid yields by supplementing with acetate. Appl Microbiol Biotechnol 23(5):330–335. https://doi.org/10.1007/BF00257028
Penning H, Conrad R (2006) Effect of inhibition of acetoclastic methanogenesis on growth of archaeal populations in an anoxic model environment. Appl Environ Microbiol 72(1):178–184
Poehlein A, Bremekamp R, Lutz VT, Schulz LM, Daniel R (2018) Draft genome sequence of the butanoic acid-producing bacterium Clostridium luticellarii DSM 29923, used for strong aromatic Chinese liquor production. Genome Announc 6(18):e00377–e00318. https://doi.org/10.1128/genomeA.00377-18
Raes SM, Jourdin L, Buisman CJ, Strik DP (2017) Continuous long-term bioelectrochemical chain elongation to butyrate. ChemElectroChem 4(2):386–395. https://doi.org/10.1002/celc.201600587
Sparling R, Daniels L (1987) The specificity of growth inhibition of methanogenic bacteria by bromoethanesulfonate. Can J Microbiol 33(12):1132–1136. https://doi.org/10.1139/m87-199
Spirito CM, Richter H, Rabaey K, Stams AJM, Angenent LT (2014) Chain elongation in anaerobic reactor microbiomes to recover resources from waste. Curr Opin Biotechnol 27:115–122. https://doi.org/10.1016/j.copbio.2014.01.003
Stieb M, Schink B (1989) Anaerobic degradation of isobutyrate by methanogenic enrichment cultures and by a Desulfococcus multivorans strain. Arch Microbiol 151(2):126–132. https://doi.org/10.1007/Bf00414426
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85(3):612–626. https://doi.org/10.1111/1574-6941.12148
Tholozan J-L, Samain E, Grivet J-P (1988) Isomerization between n-butyrate and isobutyrate in enrichment cultures. FEMS Microbiol Ecol 4(3–4):187–191. https://doi.org/10.1111/j.1574-6968.1988.tb02663.x
van der Meijden P, van der Drift C, Vogels GD (1984) Methanol conversion in Eubacterium limosum. Arch Microbiol 138(4):360–364. https://doi.org/10.1007/BF00410904
Vassilev I, Hernandez PA, Batlle-Vilanova P, Freguia S, Krömer JO, Keller JR, Ledezma P, Virdis B (2018) Microbial electrosynthesis of isobutyric, butyric, caproic acids, and corresponding alcohols from carbon dioxide. ACS Sustain Chem Eng 6(7):8485–8493. https://doi.org/10.1021/acssuschemeng.8b00739
Vilchez-Vargas R, Geffers R, Suárez-Diez M, Conte I, Waliczek A, Kaser VS, Kralova M, Junca H, Pieper DH (2013) Analysis of the microbial gene landscape and transcriptome for aromatic pollutants and alkane degradation using a novel internally calibrated microarray system. Environ Microbiol 15(4):1016–1039. https://doi.org/10.1111/j.1462-2920.2012.02752.x
Vrijbloed JW, Zerbe-Burkhardt K, Ratnatilleke A, Grubelnik-Leiser A, Robinson JA (1999) Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: influence on polyketide antibiotic biosynthesis. J Bacteriol 181(18):5600–5605
Wang C-d, Chen Q, Wang Q, Li C-H, Leng Y-Y, S-g L, Zhou X-W, Han W-J, J-g L, Zhang X-H (2014) Long-term batch brewing accumulates adaptive microbes, which comprehensively produce more flavorful Chinese liquors. Food Res Int 62:894–901. https://doi.org/10.1016/j.foodres.2014.05.017
Wang Q, Wang C-d, C-h L, J-g L, Chen Q, Y-z L (2015) Clostridium luticellarii sp. nov., isolated from a mud cellar used for producing strong aromatic liquors. Int J Syst Evol Microbiol 65(12):4730–4733. https://doi.org/10.1099/ijsem.0.000641
Wilbanks B, Trinh CT (2017) Comprehensive characterization of toxicity of fermentative metabolites on microbial growth. Biotechnol Biofuels 10(1):262. https://doi.org/10.1186/s13068-017-0952-4
Wu W-M, Jain MK, De Macario EC, Thiele JH, Zeikus JG (1992) Microbial composition and characterization of prevalent methanogens and acetogens isolated from syntrophic methanogenic granules. Appl Microbiol Biotechnol 38(2):282–290. https://doi.org/10.1007/Bf00174484
Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:17. https://doi.org/10.1093/nar/25.17.3389
Zhang K, Woodruff AP, Xiong M, Zhou J, Dhande YK (2011) A synthetic metabolic pathway for production of the platform chemical isobutyric acid. ChemSusChem 4(8):1068–1070. https://doi.org/10.1002/cssc.201100045
Zhu X, Tao Y, Liang C, Li X, Wei N, Zhang W, Zhou Y, Yang Y, Bo T (2015) The synthesis of n-caproate from lactate: a new efficient process for medium-chain carboxylates production. Sci Rep 5:14360. https://doi.org/10.1038/srep14360
Acknowledgments
The authors would like to thank Jose Maria Carvajal Arroyo and Pieter Candry for their insightful discussion, and Tim Lacoere, Ruben Props, Jan Arends, and Frederiek-Maarten Kerckhof for their suggestions on data analysis.
Funding
S.H. is supported by European Union’s Horizon 2020 research and innovation program SuPER-W under the Marie Skłodowska-Curie grant agreement no 676070. R.G. gratefully acknowledges support from the Ghent University BOF fellowship (BOF15/PDO/068). K.R. is supported by European Union’s Horizon 2020 research and innovation program PERCAL under grant agreement no 745828.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection and analysis were performed by Shengle Huang. The first draft of the manuscript was written by Shengle Huang and all authors commented on subsequent versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 666 kb)
Rights and permissions
About this article
Cite this article
Huang, S., Kleerebezem, R., Rabaey, K. et al. Open microbiome dominated by Clostridium and Eubacterium converts methanol into i-butyrate and n-butyrate. Appl Microbiol Biotechnol 104, 5119–5131 (2020). https://doi.org/10.1007/s00253-020-10551-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10551-w