Abstract
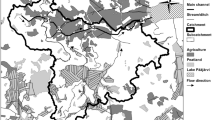
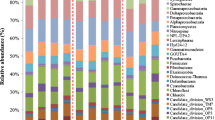
Streambank legacy sediments may be important sources of sediment and nutrients from Mid-Atlantic watersheds. However, little is known about the nutrient processing roles of microorganisms that inhabit legacy sediments, let alone their composition, diversity, and distributions. In this study, we sampled 15 streambanks at multiple depths throughout four watersheds in the Mid-Atlantic Region of the USA. High throughput sequencing of 16S ribosomal RNA genes indicated that streambank microbial community composition varied within site depth and across contemporary land uses. Collectively, the most abundant microbial taxa in legacy sediments included Acidobacteria (25–45%), Proteobacteria (15–40%), Nitrospirae (2–10%), Chloroflexi (1–5%), and Actinobacteria (1–10%). Bacterial community composition was distinct between agriculture and urban sites as well as suburban and urban sites. There was significant variation in community composition between the top (1–25%), upper-middle (26–50%), and bottom layers (76–100%) of sediments, while the relative abundances differed between layers for only Acidobacteria and Proteobacteria. Several streambank chemistry variables (metals, %TC, and %TN) had weak positive correlations with community composition. Compared to ammonia-oxidizing bacteria, nitrifying archaea were more predominant. This study provides the first insights into detailed microbial composition of legacy sediments and identifies environmental drivers for community structure and nitrogen processing. Future studies should consider exploring the role of this unique microbial environment for nutrient processing and leaching from legacy sediments and its implications for watershed water quality.






Similar content being viewed by others
References
Alekseyenko AV (2016) Multivariate welch t-test on distances. Bioinformatics 32(23):3552–3558. https://doi.org/10.1093/bioinformatics/btw524
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Aust J Ecol 26:32–46. https://doi.org/10.1046/j.1442-9993.2001.01070.x
Argiroff WA, Zak DR, Lanser CM, Wiley MJ (2017) Microbial community functional potential and composition are shaped by hydrologic connectivity in riverine floodplain soils. Microb Ecol 73:630–644. https://doi.org/10.1007/s00248-016-0883-9
Banks WSL, Gellis AC, Noe G (2010) Sources of fine-grained suspended sediment in Mill Stream Branch Watershed, Corsica River Basin, a tributary to the Chesapeake Bay, Maryland, 2009. In: Proceedings, 2nd joint federal interagency conference, Las Vegas, NV, June 27–July 1, 2010. CD-ROM ISBN 978-0-0779007-3-2, 6B, pp 1–12
Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N (2011) The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43:1450–1455. https://doi.org/10.1016/j.soilbio.2011.03.012
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr 27:325–349
Brewer TE, Aronson EL, Arogyaswamy K, Billings SA, Botthoff JK, Campbell AN, Dove NC, Fairbanks D, Gallery RE, Hart SC, Kaye J, King G, Logan G, Lohse KA, Maltz MR, Mayorga E, O’Neill C, Owens SM, Packman A, Pett-Ridge J, Plante AF, Richter DD, Silver WL, Yang WH, Fierer N (2019) Ecological and genomic attributes of novel bacterial taxa that thrive in subsurface soil horizons. MBio 10:1–14. https://doi.org/10.1128/mBio.01318-19
Buckley DH, Schmidt TM (2003) Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ Microbiol 5:441–452
Callahan B (2016) The RDP and GreenGenes taxonomic training sets formatted for DADA2 [Data Set]. Zenodo. https://doi.org/10.5281/zenodo.158955
Carini P, Delgado-Baquerizo M, Hinckley E-LS, Brewer TE, Garrett R, Vanderburgh C, McKnight D, Fierer N (2018) Unraveling the effects of spatial variability and relic DNA on the temporal dynamics of soil microbial communities 1–19. https://doi.org/10.1101/402438
Cashman MJ, Gellis A, Sanisaca LG, Noe GB, Cogliandro V, Baker A (2018) Bank-derived material dominates fluvial sediment in a suburban Chesapeake Bay watershed. River Res Appl. https://doi.org/10.1002/rra.3325
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Childers SE, Ciufo S, Lovley DR (2002) Geobacter metallireducens accesses insoluble Fe( III ) oxide by chemotaxis. Nature 416:767–769
Clarke KR (1993) Nonparametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Crowther TW, Maynard DS, Leff JW (2014) Predicting the responsiveness of soil biodiversity to deforestation: a cross-biome study. Glob Change Biol. https://doi.org/10.1111/gcb.12565
Delaware Department of Natural Resources and Environmental Control (DNREC) (2011) Christina Basin pollution control strategy. https://www.dnrec.delaware.gov/swc/wa/Pages/ChristinaBasin.aspx
Dodbiba G, Ponou J, Fujita T (2015) Biosorption of heavy metals. In: Abhilash Pandey BD, Natarajan KA (eds) Materials for minerals, metals and materials and the environment. CRC Press, Boca Raton, pp 409–426
Dunbar J, Barns SM, Ticknor LO, Kuske CR (2002) Empirical and theoretical bacterial diversity in four Arizona soils. Appl Environ Microbiol 68:3035–3045. https://doi.org/10.1128/AEM.68.6.3035
Duval TP, Hill AR (2007) Influence of base flow stream bank seepage on riparian zone N biogeochemistry. Biogeochemistry 85:185–199. https://doi.org/10.1007/s10533-007-9128-9
Einen J, Thorseth IH, Øvreås L (2008) Enumeration of Archaea and Bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy. FEMS Microbiol Lett 282:182–187. https://doi.org/10.1111/j.1574-6968.2008.01119.x
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579–590. https://doi.org/10.1038/nrmicro.2017.87
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci 103:626–631. https://doi.org/10.1073/pnas.0507535103
Fierer N, Bradford M, Jackson R (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, Knight R, Gilbert JA, McCulley RL (2013) Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 342:621–624. https://doi.org/10.1126/science.1243768
Forshay KJ, Stanley EH (2005) Rapid nitrate loss and denitrification in a temperate river floodplain. Biogeochemistry 75:43–64. https://doi.org/10.1007/s10533-004-6016-4
Fox GA, Purvis RA, Penn CJ (2016) Streambanks: a net source of sediment and phosphorus to streams and rivers. J Environ Manage 181:602–614. https://doi.org/10.1016/j.jenvman.2016.06.071
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. PNAS 102:14683–14688
Gellis AC, Mukundan R (2013) Watershed sediment source identification: tools, approaches, and case studies. J Soils Sediments 13:1655–1657. https://doi.org/10.1007/s11368-013-0778-z
Gellis AC, Noe GB (2013) Sediment source analysis in the Linganore Creek watershed, Maryland, USA, using the sediment fingerprinting approach: 2008 to 2010. J Soils Sediments 13:1735–1753. https://doi.org/10.1007/s11368-013-0771-6
Gellis AC, Hupp CR, Pavich MJ, Landwehr JM, Banks WSL, Hubbard BE, Langland MJ, Ritchie JC, Reuter JM (2009) Sources, transport, and storage of sediment at selected sites in the Chesapeake Bay Watershed. US Environmental Protection Agency Chesapeake Bay Program, New York
Gellis AC, Myers MK, Noe GB, Hupp CR, Schenk ER, Myers L (2017) Storms, channel changes, and a sediment budget for an urban-suburban stream, difficult run, Virginia, USA. Geomorphology 278:128–148. https://doi.org/10.1016/j.geomorph.2016.10.031
Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, Yannarell A et al (2016) Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Frontiers Microbiol 7:1–10. https://doi.org/10.3389/fmicb.2016.00214
Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13:1642–1654. https://doi.org/10.1111/j.1462-2920.2011.02480.x
Gurwick NP, Groffman PM, Yavitt JB, Gold AJ, Blazejewski G, Stolt M (2008a) Microbially available carbon in buried riparian soils in a glaciated landscape. Soil Biol Biochem 40:85–96. https://doi.org/10.1016/j.soilbio.2007.07.007
Gurwick NP, Mccorkle DM, Groffman PM, Gold AJ, Kellogg DQ, Seitz-rundlett P (2008b) Mineralization of ancient carbon in the subsurface of riparian forests. J Geophys Res 113:1–13. https://doi.org/10.1029/2007JG000482
Hamidi B, Wallace K, Vasu C, Alekseyenko AV (2019) Wd-test: robust distance-based multivariate analysis of variance. Microbiome 7:51. https://doi.org/10.1186/s40168-019-0659-9
Hallin S, Philippot L, Löf FE, Sanford RA, Jones CM (2018) Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol 26:43–55. https://doi.org/10.1016/j.tim.2017.07.003
Hanson RB, Wiebe WJ (1977) Heterotrophic activity associated with particulate size fractions in a Spartina alterniflora salt-marsh estuary, Sapelo Island, Georgia, USA, and the continental shelf waters. Mar Biol 42:321–330. https://doi.org/10.1007/BF00402194
Hedin LO, von Fischer JC, Ostrom NE, Kennedy BP, Brown MG, Robertson GP (1998) Thermodynamic constraints on nitrogen transformations and other biogeochemical processes at soil–stream interfaces. Ecology 79:684–703. https://doi.org/10.1890/0012-9658(1998)079[0684:TCONAO]2.0.CO;2
Herrmann M, Scheibe A, Avrahami S, Küsel K (2011) Ammonium availability affects the ratio of ammonia-oxidizing bacteria to ammonia-oxidizing archaea in simulated creek ecosystems. Appl Environ Microbiol 77:1896–1899. https://doi.org/10.1128/AEM.02879-10
Hill A, Cardaci M (2004) Denitrification and organic carbon availability in riparian wetland soils and subsurface sediments. Soil Sci Soc Am J. https://doi.org/10.2136/sssaj2004.0320
Hou Z, Nelson WC, Stegen JC, Murray CJ, Arntzen E, Crump AR, Kennedy DW, Perkins MC, Scheibe TD, Fredrickson JK, Zachara JM (2017) Geochemical and microbial community attributes in relation to hyporheic zone geological facies. Sci Rep 7:1–17. https://doi.org/10.1038/s41598-017-12275-w
Hunter JD (2007) Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95
James LA (2013) Legacy sediment: definitions and processes of episodically produced anthropogenic sediment. Anthropocene. https://doi.org/10.1016/j.ancene.2013.04.001
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. https://doi.org/10.1128/AEM.72.3.1719
Jiang G, Lutgen A, Mattern K, Sienkiewicz N, Kan J, Inamdar S (2020) Streambank legacy sediment contributions to suspended sediment and nutrient yields from a Mid-Atlantic, Piedmont Watershed (In Review). Journal of American Water Resources Association.
Johnson C, Albrecht G, Ketterings Q, Beckman J, Stockin K (2005) Nitrogen basics—the nitrogen cycle. Cornell University Cooperative Extension, pp 1–3
Jones SE, Lennon JT (2010) Dormancy contributes to the maintenance of microbial diversity. PNAS 107:5881–5886. https://doi.org/10.1073/pnas.0912765107
Kan J (2018) Storm events restructured bacterial community and their biogeochemical potentials. J Geophys Res Biogeosci 123:2257–2269. https://doi.org/10.1029/2017JG004289
Kaushal SS, Groffman PM, Mayer PM, Striz E, Gold AJ (2008) Effects of stream restoration on denitrification in an urbanizing watershed. Ecol Appl 18:789–804
Kielak AM, Barreto CC, Kowalchuk GA, van Veen JA, Kuramae EE (2016) The ecology of acidobacteria: moving beyond genes and genomes. Frontiers Microbiol 7:1–16. https://doi.org/10.3389/fmicb.2016.00744
Kim H, Bae HS, Reddy KR, Ogram A (2016) Distributions, abundances and activities of microbes associated with the nitrogen cycle in riparian and stream sediments of a river tributary. Water Res 106:51–61. https://doi.org/10.1016/j.watres.2016.09.048
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. https://doi.org/10.1093/nar/gks808
Kloos K, Mergel A, Rosch C, Bothe H (2001) Denitrification within the genus Azospirillum and other associative bacteria. Funct Plant Biol 28:991–998
Koval J (2011) Assessing restoration potential in relict wetland soils: investigating the effect of wetland hydrology on soil microbial community composition and denitrification potential. MS Thesis. University of Illinois, Urbana-Champaign.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. https://doi.org/10.1038/nature04983
Li D, Sharp JO, Drewes JE (2016) Influence of wastewater discharge on the metabolic potential of the microbial community in river sediments. Microb Ecol 71:78–86. https://doi.org/10.1007/s00248-015-0680-x
Liu Y, Liu Y, Ding Y, Zheng J, Zhou T, Pan G, Crowley D, Li L, Zheng J, Zhang X, Yu X, Wang J, Reigosa M (2014) Abundance, composition and activity of ammonia oxidizer and denitrifier communities in metal polluted rice paddies from South China. PLoS ONE 9(7):e102000
Lutgen A (2019) Legacy sediment stream bank chemical content: variations with land use, bank depth, and season. MS Thesis. University of Delaware.
Lutgen A, Jiang G, Sienkiewicz N, Mattern K, Kan J, Inamdar S (2020) Variations in nutrient and heavy metal concentrations for streambank legacy sediments and their implications for water quality (In review). J Am Water Resources Assoc.
Mayer PM, Groffman PM, Striz EA, Kaushal SS (2010) Nitrogen dynamics at the groundwater-surface water interface of a degraded urban stream. J Environ Qual 39(3):810–823
Maynard W (2015) An intimate portrait. University of Pennsylvania Press, Philadelphia
Mehlich A (1984) Mehlich 3 soil test extractant in: a modification of Mehlich 2 extractant. Commun Soil Sci Plant Anal 15(12):1409
Merritts D, Walter R, Rahnis M, Hartranft J, Cox S, Gellis A et al (2011) Anthropocene streams and base-level controls from historic dams in the unglaciated mid-Atlantic region, USA. Philosoph Trans Math Phys Eng Sci 369:976–1009. https://doi.org/10.1098/rsta.2010.0335
Merritts D, Walter R, Cox S, Potter N, Kratz L (2013) The rise and fall of Mid-Atlantic streams: millpond sedimentation, milldam breaching, channel incision, and stream bank erosion. Geol Soc Am 4121:183–203. https://doi.org/10.1130/2013.4121(14)
Miller A, Baker M, Boomer K, Merritts D, Prestegaard K, Smith S (2019) Legacy sediment, riparian corridors, and total maximum daily loads. STAC workshop report annapolis, MD
Niemitz J, Haynes C, Lasher G (2013) Legacy sediments and historic land use: chemostratigraphic evidence for excess nutrient and heavy metal sources and remobilization. Geology 41:47–50. https://doi.org/10.1130/G33547.1
Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlin D, et al (2018) vegan: Community ecology package. R package version 2.5-2. https://rdrr.io/cran/vegan/
Rocca JD, Hall EK, Lennon JT, Evans SE, Waldrop MP, Cotner JB, Nemergut DR, Graham EB, Wallenstein MD (2014) Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J 9:1693–1699. https://doi.org/10.1038/ismej.2014.252
Sanford RA, Wagner DD, Wu Q, Chee-sanford JC, Thomas SH, Cruz-garcía C, Rodríguez G, Massol-deyá A, Krishnani KK, Ritalahti KM (2012) Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. PNAS 109:19709–19714. https://doi.org/10.1073/pnas.1211238109
Serna-chavez HM, Fierer N, Van Bodegom PM (2013) Global drivers and patterns of microbial abundance in soil. Glob Ecol Biogeogr 22:1162–1172. https://doi.org/10.1111/geb.12070
Straub KL (2011) Geobacter, 1 edn. Encyclopedia of Earth Sciences Series. Elsevier Ltd., Amsterdam. https://doi.org/10.1007/978-1-4020-9212-1_99
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712. https://doi.org/10.1128/AEM.NA
Sims JT, Gartley KL, Pautler MC, Leytem AB, Maguire RO (2002) Evaluation of Mehlich 3 as an agri-environmental soil phosphorus test for the mid-Atlantic United States of America. Soil Sci Soc Am J 66:2016–2032. https://doi.org/10.2136/sssaj2002.2016
Strickland MS, Lauber C, Fierer N, Bradford MA (2009) Testing the functional significance of microbial community composition. Ecology 90:441–451
Throbäck IN, Enwall K, Jarvis Å, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49:401–417. https://doi.org/10.1016/j.femsec.2004.04.011
Walter RC, Merritts DJ (2008) Natural streams of water-powered and the legacy mills. Am Assoc Adv Sci 319:299–304
Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M et al (2009) Three genomes from the phylum acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75:2046–2056. https://doi.org/10.1128/AEM.02294-08
Waskom M, Botvinnik O, O'Kane D, Hobson P, Ostblom J, Lukauskas S et al (2018) mwaskom/seaborn: v0.9.0 (July 2018) (Version v0.9.0). Zenodo. https://doi.org/10.5281/zenodo.1313201
Webster G, Embley TM, Freitag TE, Smith Z, Prosser JI (2005) Links between ammonia oxidizer species composition, functional diversity and nitrification kinetics in grassland soils. Environ Microbiol 7:676–684. https://doi.org/10.1111/j.1462-2920.2005.00740.x
Wegmann KW, Lewis RQ, Hunt MC (2012) Historic mill ponds and piedmont stream water quality: making the connection near Raleigh, North Carolina. The Geological Society of America 29:93–121. https://doi.org/10.1130/2012.0029(03)
Wegmann KW, Osburn CL, Lewis RQ, Peszlen IM, Mitasova H (2013) Legacy sediments and stream water quality: estimating volume, nutrient content, and stream bank erosion in 303(d)-impaired waterways of the North Carolina Piedmont. Report N. 435, Water Resources Research Institute of the University of North Carolina.
Weisstein EW (2004) Bonferroni correction. Wolfram Research. https://mathworld.wolfram.com/BonferroniCorrection.html
Weitzman JN, Kaye JP (2017) Nitrate retention capacity of milldam-impacted legacy sediments and relict A horizon soils. SOIL 3:95–112. https://doi.org/10.5194/soil-3-95-2017
Weitzman JN, Forshay KJ, Kaye JP, Mayer PM, Koval JC, Walter RC (2014) Potential nitrogen and carbon processing in a landscape rich in milldam legacy sediments. Biogeochemistry 120:337–357. https://doi.org/10.1007/s10533-014-0003-1
Wolman MG (1959) Factors influencing erosion of a cohesive river bank. Am J Sci 257:204–216
Zhang X, Gu Q, Long X, Li Z, Liu D, Ye D, He C-Q, Liu X-Y, Vaananen K, Chen X-P (2016) Anthropogenic activities drive the microbial community and its function in urban river sediment. J Soils Sediments 16:716–725. https://doi.org/10.1007/s11368-015-1246-8
Acknowledgements
Funding for this project was provided by the US Department of Agriculture [Grant Number USDA-NIFA 2017-67019-26330], William Penn Foundation [Grant Number 188-17], and Endowment Fund of Stroud Water Research Center. We thank the Fair Hill Natural Resources Management Area staff, the Stroud Water Research Center staff, the University of Delaware Soils Testing Lab, and the University of Maryland Center for Environmental Science for their support. We express our gratitude to Drs. Dorothy Merritts and Robert Walter for their knowledge and field insights that they generously provided for this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Jennifer Leah Tank.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sienkiewicz, N., Bier, R.L., Wang, J. et al. Bacterial communities and nitrogen transformation genes in streambank legacy sediments and implications for biogeochemical processing. Biogeochemistry 148, 271–290 (2020). https://doi.org/10.1007/s10533-020-00659-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10533-020-00659-6