Oligomerization and Conformational Change Turn Monomeric β-Amyloid and Tau Proteins Toxic: Their Role in Alzheimer’s Pathogenesis
Abstract
:1. Introduction
- The acetylcholine hypothesis: loss of cholinergic neurons and neurotransmission causes AD [2].
- The tau hypothesis [5]: abnormal phosphorylation of tau proteins is in the background of AD progress.
- The dual cascade hypothesis: cellular processes simultaneously drive tau and Aβ pathology in the cortex [6].
- The mitochondrial dysfunction hypothesis [7].
- The neuroinflammation hypothesis [8] emphasizes the role of immune attack in neuronal loss.
- The vascular dysfunction hypothesis [9] underlines the role of brain circulation and endothelial-mediated processes in AD pathogenesis.
- Peripheral Aβ hypothesis: β-amyloid peptides enter the brain from the blood from the periphery [10].
- The cellular hypothesis [1] represents a complex view of AD pathogenesis, which emphasizes the central role of aging as the major driver of the disease and the participation of different brain cells (microglia, astrocytes, and brain vasculature) in the pathophysiology.
2. Amyloid Peptides
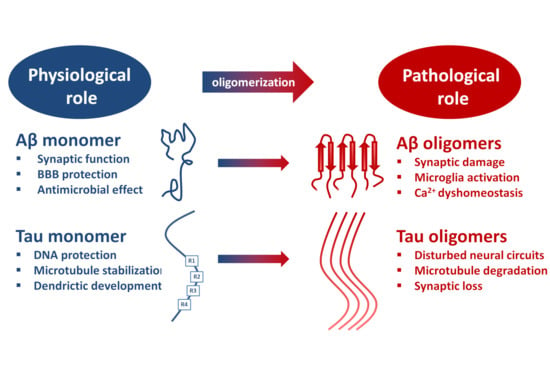
2.1. The Physiological Role of β-Amyloid Precursor Protein (APP) and the Monomeric Aβ Peptide
- Regulation of synaptic function by activating nicotinic ACh receptors and participation in memory consolidation. Aβ monomer homeostasis is essential for normal synaptic function [38].
- Promotion of recovery after traumatic brain injury.
- Protection of the blood-brain barrier by blocking leaks and preventing leakage.
- Antimicrobial activity, probably by disturbing membrane structures of microbes.
- Suppression of tumor growth through inhibition of viruses.
2.2. Formation of AβOs and Fibrils by Aggregation
|
|
|
|
|
|
|
|
|
|
2.3. The Structure of Aβ Species
2.3.1. Structure of Aβ Monomers and Oligomers
2.3.2. Structure of Fibrillar Aβ (fAβ)
2.4. Mechanisms of Toxicity Induced by AβOs
- The membrane hypothesis focuses on the lipid membrane–AβO interactions. AβOs can deteriorate membrane structures, insert directly into cell membranes, and may form pores [104] inducing Ca-influx to the cells [104]. The ganglioside GM1 plays a decisive role in the membrane–AβO interactions and AβO toxicity [105].
- According to the receptor hypothesis, AβOs are pathogenic ligands that can bind to specific receptors [106,107,108,109]. Several facts support this hypothesis:
- saturation of the binding sites,
- binding specificity for definite neural cells,
- targeting of synapses,
- receptor antagonists inhibit the toxic effect,
- the binding sites are trypsin sensitive proteins
- Cellular prion protein (PrPc)
- Na,K-ATPase alpha 3 subunit (NKAa3)
- RAGE receptor
- Tyrosine kinase ephrin type A receptor
- Tyrosine kinase ephrin type B receptor
- p75 neurotrophin receptor
- leucocyte immunoglobulin receptor B2 (LILRB2)
- triggering receptor expressed on myeloid cells 2 (TREM2)
- toll-like receptor 2 (TLR2R)
- neuroligin
- NMDA receptors
- AMPA receptors
- metabotropic Glu receptor 5
- frizzled receptor [112]
- alpha7nACh receptor
- adrenergic receptors
- calcium channels
- N-formyl peptide receptor 2
- IgGFc region receptor II b
- Transient receptor potential melastatin (TRPM2)
- insulin receptor
- Wnt receptor
2.5. Intracellular Aβ and Extracellular Plaques
3. Tau Proteins
3.1. Tau Isoforms, Domain Structure, Post-Translational Modifications
3.2. Physiological Actions of Tau
3.3. The Structure of Tau Oligomers and Fibrils
3.4. Pathophysiological Effects of Tau Assemblies.
4. Genetic Risk Factors of AD: Classical Studies and Novel Results
- a)
- b)
- ApoE also modulates tau pathology; especially ApoE4 enhances tau pathogenicity.
- c)
- ApoE4 mediates AD risk by modulation of immune and microglial responses to amyloid plaques.
- d)
- ApoE4 has direct pathological effects on neurons and neuronal networks. ApoE3 expressions in neurons are protective by stimulating neurite outgrowth.
- e)
- ApoE4 directly impairs the blood-brain-barrier function in AD.
5. Aβ-Tau Crosstalk
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- De Strooper, B.; Karran, E. The Cellular Phase of Alzheimer’s Disease. Cell 2016, 164, 603–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, P.T.; Palmer, A.M.; Snape, M.; Wilcock, G.K. The cholinergic hypothesis of Alzheimer’s disease: A review of progress. J. Neurol. Neurosurg. Psychiatry 1999, 66, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Allsop, D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991, 12, 383–388. [Google Scholar] [CrossRef]
- Lambert, M.P.; Barlow, A.K.; Chromy, B.A.; Edwards, C.; Freed, R.; Liosatos, M.; Morgan, T.E.; Rozovsky, I.; Trommer, B.; Viola, K.L.; et al. Diffusible, nonfibrillar ligands derived from A 1-42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. 1998, 95, 6448–6453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundke-Iqbal, I.; Iqbal, K.; Tung, Y.C.; Quinlan, M.; Wisniewski, H.M.; Binder, L.I. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. 1986, 83, 4913–4917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Small, S.A.; Duff, K. Linking Aβ and Tau in Late-Onset Alzheimer’s Disease: A Dual Pathway Hypothesis. Neuron 2008, 60, 534–542. [Google Scholar] [CrossRef] [Green Version]
- Swerdlow, R.H.; Burns, J.M.; Khan, S.M. The Alzheimer’s disease mitochondrial cascade hypothesis: Progress and perspectives. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 2014, 1842, 1219–1231. [Google Scholar] [CrossRef] [Green Version]
- Heppner, F.L.; Ransohoff, R.M.; Becher, B. Immune attack: The role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 2015, 16, 358–372. [Google Scholar] [CrossRef]
- Di Marco, L.Y.; Venneri, A.; Farkas, E.; Evans, P.C.; Marzo, A.; Frangi, A.F. Vascular dysfunction in the pathogenesis of Alzheimer’s disease — A review of endothelium-mediated mechanisms and ensuing vicious circles. Neurobiol. Dis. 2015, 82, 593–606. [Google Scholar] [CrossRef]
- Bu, X.-L.; Xiang, Y.; Jin, W.-S.; Wang, J.; Shen, L.-L.; Huang, Z.-L.; Zhang, K.; Liu, Y.-H.; Zeng, F.; Liu, J.-H.; et al. Blood-derived amyloid-β protein induces Alzheimer’s disease pathologies. Mol. Psychiatry 2018, 23, 1948–1956. [Google Scholar] [CrossRef]
- Kovacs, G. Molecular Pathological Classification of Neurodegenerative Diseases: Turning towards Precision Medicine. Int. J. Mol. Sci. 2016, 17, 189. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, J.; Kovacs, G.G. Prevalence of mixed pathologies in the aging brain. Alzheimers Res. Ther. 2014, 6, 82–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duyckaerts, C.; Delatour, B.; Potier, M.-C. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 2009, 118, 5–36. [Google Scholar] [CrossRef] [PubMed]
- Castellani, R.J.; Rolston, R.K.; Smith, M.A. Alzheimer Disease. Dis. Mon. 2010, 56, 484–546. [Google Scholar] [CrossRef]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef]
- Choi, M.L.; Gandhi, S. Crucial role of protein oligomerization in the pathogenesis of Alzheimer’s and Parkinson’s diseases. FEBS J. 2018, 285, 3631–3644. [Google Scholar] [CrossRef] [Green Version]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef]
- Cline, E.N.; Bicca, M.A.; Viola, K.L.; Klein, W.L. The Amyloid-β Oligomer Hypothesis: Beginning of the Third Decade. J. Alzheimers Dis. 2018, 64, S567–S610. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-Q.; Mobley, W.C. Alzheimer Disease Pathogenesis: Insights From Molecular and Cellular Biology Studies of Oligomeric Aβ and Tau Species. Front. Neurosci. 2019, 13, 659–680. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, A.B. Progress Report on Alzheimer’s Disease 2004–2005; U.S. Department of Health and Human Services, National Institutes on Aging, National Institutes of Health: Washington, DC, USA, 2005. Available online: http://www.agis.com/SqlFileResource.axd?id=441&resource=pdf (accessed on 27 March 2020).
- Castanho, I.; Murray, T.K.; Hannon, E.; Jeffries, A.; Walker, E.; Laing, E.; Baulf, H.; Harvey, J.; Bradshaw, L.; Randall, A.; et al. Transcriptional Signatures of Tau and Amyloid Neuropathology. Cell Rep. 2020, 30, 2040–2054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panza, F.; Lozupone, M.; Logroscino, G.; Imbimbo, B.P. A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2019, 15, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Penke, B.; Bogár, F.; Paragi, G.; Gera, J.; Fülöp, L. Key Peptides and Proteins in Alzheimer’s Disease. Curr. Protein Pept. Sci. 2019, 20, 577–599. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, E.; Small, D.H. Insights into the physiological function of the β-amyloid precursor protein: Beyond Alzheimer’s disease. J. Neurochem. 2014, 129, 756–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nhan, H.S.; Chiang, K.; Koo, E.H. The multifaceted nature of amyloid precursor protein and its proteolytic fragments: Friends and foes. Acta Neuropathol. 2015, 129, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldwell, J.H.; Klevanski, M.; Saar, M.; Müller, U.C. Roles of the amyloid precursor protein family in the peripheral nervous system. Mech. Dev. 2013, 130, 433–446. [Google Scholar] [CrossRef]
- Zhou, R.; Yang, G.; Guo, X.; Zhou, Q.; Lei, J.; Shi, Y. Recognition of the amyloid precursor protein by human γ-secretase. Science 2019, 363, eaaw0930. [Google Scholar] [CrossRef]
- Tam, J.H.; Seah, C.; Pasternak, S.H. The Amyloid Precursor Protein is rapidly transported from the Golgi apparatus to the lysosome and where it is processed into beta-amyloid. Mol. Brain 2014, 7, 54–72. [Google Scholar] [CrossRef] [Green Version]
- Deyts, C.; Thinakaran, G.; Parent, A.T. APP Receptor? To Be or Not To Be. Trends Pharmacol. Sci. 2016, 37, 390–411. [Google Scholar] [CrossRef] [Green Version]
- Wildburger, N.C.; Esparza, T.J.; LeDuc, R.D.; Fellers, R.T.; Thomas, P.M.; Cairns, N.J.; Kelleher, N.L.; Bateman, R.J.; Brody, D.L. Diversity of Amyloid-beta Proteoforms in the Alzheimer’s Disease Brain. Sci. Rep. 2017, 7, 9520–9529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudinskiy, N.; Fuerer, C.; Demurtas, D.; Zamorano, S.; De Piano, C.; Herrmann, A.G.; Spires-Jones, T.L.; Oeckl, P.; Otto, M.; Frosch, M.P.; et al. Amyloid-beta oligomerization is associated with the generation of a typical peptide fragment fingerprint. Alzheimers Dement. 2016, 12, 996–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galante, D.; Ruggeri, F.S.; Dietler, G.; Pellistri, F.; Gatta, E.; Corsaro, A.; Florio, T.; Perico, A.; D’Arrigo, C. A critical concentration of N-terminal pyroglutamylated amyloid beta drives the misfolding of Ab1-42 into more toxic aggregates. Int. J. Biochem. Cell Biol. 2016, 79, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Tharp, W.G.; Sarkar, I. Origins of amyloid-β. BMC Genomics 2013, 14, 290–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brothers, H.M.; Gosztyla, M.L.; Robinson, S.R. The Physiological Roles of Amyloid-β Peptide Hint at New Ways to Treat Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 118–134. [Google Scholar] [CrossRef]
- Wang, S.; Bolós, M.; Clark, R.; Cullen, C.L.; Southam, K.A.; Foa, L.; Dickson, T.C.; Young, K.M. Amyloid β precursor protein regulates neuron survival and maturation in the adult mouse brain. Mol. Cell. Neurosci. 2016, 77, 21–33. [Google Scholar] [CrossRef]
- Hillen, H. The Beta Amyloid Dysfunction (BAD) Hypothesis for Alzheimer’s Disease. Front. Neurosci. 2019, 13, 1154–1164. [Google Scholar] [CrossRef] [Green Version]
- Morley, J.E.; Farr, S.A. Hormesis and Amyloid-β Protein: Physiology or Pathology? J. Alzheimers Dis. 2012, 29, 487–492. [Google Scholar] [CrossRef]
- Lutter, L.; Serpell, C.J.; Tuite, M.F.; Xue, W.-F. The molecular lifecycle of amyloid – Mechanism of assembly, mesoscopic organisation, polymorphism, suprastructures, and biological consequences. Biochim. Biophys. BBA - Proteins Proteomics 2019, 1867, 140257–140271. [Google Scholar] [CrossRef]
- Fändrich, M. Oligomeric Intermediates in Amyloid Formation: Structure Determination and Mechanisms of Toxicity. J. Mol. Biol. 2012, 421, 427–440. [Google Scholar] [CrossRef]
- Ferreira, S.T.; Lourenco, M.V.; Oliveira, M.M.; De Felice, F.G. Soluble amyloid-β oligomers as synaptotoxins leading to cognitive impairment in Alzheimer’s disease. Front. Cell. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, W.; Wang, Z.; Liu, W.; O’Malley, T.T.; Jin, M.; Willem, M.; Haass, C.; Frosch, M.P.; Walsh, D.M. Diffusible, highly bioactive oligomers represent a critical minority of soluble Aβ in Alzheimer’s disease brain. Acta Neuropathol. 2018, 136, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Georganopoulou, D.G.; Chang, L.; Nam, J.-M.; Thaxton, C.S.; Mufson, E.J.; Klein, W.L.; Mirkin, C.A. From The Cover: Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer’s disease. Proc. Natl. Acad. Sci. 2005, 102, 2273–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foroutanpay, B.V.; Kumar, J.; Kang, S.G.; Danaei, N.; Westaway, D.; Sim, V.L.; Kar, S. The Effects of N-terminal Mutations on β-amyloid Peptide Aggregation and Toxicity. Neuroscience 2018, 379, 177–188. [Google Scholar] [CrossRef]
- Thu, T.T.M.; Co, N.T.; Tu, L.A.; Li, M.S. Aggregation rate of amyloid beta peptide is controlled by beta-content in monomeric state. J. Chem. Phys. 2019, 150, 225101–225109. [Google Scholar] [CrossRef]
- Hamdan, N.; Kritsiligkou, P.; Grant, C.M. ER stress causes widespread protein aggregation and prion formation. J. Cell Biol. 2017, 216, 2295–2304. [Google Scholar] [CrossRef] [Green Version]
- Barz, B.; Strodel, B. Understanding Amyloid-β Oligomerization at the Molecular Level: The Role of the Fibril Surface. Chem. - Eur. J. 2016, 22, 8768–8772. [Google Scholar] [CrossRef]
- Yan, T.; Liang, J.; Gao, J.; Wang, L.; Fujioka, H.; Zhu, X.; Wang, X. FAM222A encodes a protein which accumulates in plaques in Alzheimer’s disease. Nat. Commun. 2020, 11, 411–427. [Google Scholar] [CrossRef]
- Galzitskaya, O.V.; Galushko, E.I.; Selivanova, O.M. Studies of the Process of Amyloid Formation by Aβ Peptide. Biochem. Mosc. 2018, 83, S62–S80. [Google Scholar] [CrossRef]
- Bitan, G.; Kirkitadze, M.D.; Lomakin, A.; Vollers, S.S.; Benedek, G.B.; Teplow, D.B. Amyloid - protein (A) assembly: A 40 and A 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. 2003, 100, 330–335. [Google Scholar] [CrossRef] [Green Version]
- Törnquist, M.; Michaels, T.C.T.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.A.; Knowles, T.P.J.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, M.C.; Gnutt, D.; Gao, M.; Wärmländer, S.K.T.S.; Jarvet, J.; Gräslund, A.; Winter, R.; Ebbinghaus, S.; Strodel, B. Effects of in vivo conditions on amyloid aggregation. Chem. Soc. Rev. 2019, 48, 3946–3996. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Shen, Y.; Lee, J.H.; Ying, J.; Bax, A. Monomeric Aβ 1–40 and Aβ 1–42 Peptides in Solution Adopt Very Similar Ramachandran Map Distributions That Closely Resemble Random Coil. Biochemistry 2016, 55, 762–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Shea, J.-E. Effects of Solvent on the Structure of the Alzheimer Amyloid-β(25–35) Peptide. Biophys. J. 2006, 91, 1638–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Tsai, M.-Y.; Wolynes, P.G. Comparing the Aggregation Free Energy Landscapes of Amyloid Beta(1–42) and Amyloid Beta(1–40). J. Am. Chem. Soc. 2017, 139, 16666–16676. [Google Scholar] [CrossRef] [Green Version]
- Dovidchenko, N.V.; Glyakina, A.V.; Selivanova, O.M.; Grigorashvili, E.I.; Suvorina, M.Y.; Dzhus, U.F.; Mikhailina, A.O.; Shiliaev, N.G.; Marchenkov, V.V.; Surin, A.K.; et al. One of the possible mechanisms of amyloid fibrils formation based on the sizes of primary and secondary folding nuclei of Aβ40 and Aβ42. J. Struct. Biol. 2016, 194, 404–414. [Google Scholar] [CrossRef]
- Ahmed, M.; Davis, J.; Aucoin, D.; Sato, T.; Ahuja, S.; Aimoto, S.; Elliott, J.I.; Van Nostrand, W.E.; Smith, S.O. Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat. Struct. Mol. Biol. 2010, 17, 561–567. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Edalji, R.; Harlan, J.E.; Holzman, T.F.; Lopez, A.P.; Labkovsky, B.; Hillen, H.; Barghorn, S.; Ebert, U.; Richardson, P.L.; et al. Structural Characterization of a Soluble Amyloid β-Peptide Oligomer. Biochemistry 2009, 48, 1870–1877. [Google Scholar] [CrossRef]
- Haupt, C.; Leppert, J.; Rönicke, R.; Meinhardt, J.; Yadav, J.K.; Ramachandran, R.; Ohlenschläger, O.; Reymann, K.G.; Görlach, M.; Fändrich, M. Structural Basis of β-Amyloid-Dependent Synaptic Dysfunctions. Angew. Chem. Int. Ed. 2012, 51, 1576–1579. [Google Scholar] [CrossRef]
- Xiao, Y.; Matsuda, I.; Inoue, M.; Sasahara, T.; Hoshi, M.; Ishii, Y. NMR-based site-resolved profiling of β-amyloid misfolding reveals structural transitions from pathologically relevant spherical oligomer to fibril. J. Biol. Chem. 2020, 295, 458–467. [Google Scholar] [CrossRef]
- Tycko, R. Amyloid Polymorphism: Structural Basis and Neurobiological Relevance. Neuron 2015, 86, 632–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meinhardt, J.; Sachse, C.; Hortschansky, P.; Grigorieff, N.; Fändrich, M. Aβ(1-40) Fibril Polymorphism Implies Diverse Interaction Patterns in Amyloid Fibrils. J. Mol. Biol. 2009, 386, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.G.; Paula-Barbosa, M.; Roher, A. Alzheimer’s disease: Paired helical filaments and cytomembranes. Neuropathol. Appl. Neurobiol. 1987, 13, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.L.; Kim, C.; Haldiman, T.; ElHag, M.; Mehndiratta, P.; Pichet, T.; Lissemore, F.; Shea, M.; Cohen, Y.; Chen, W.; et al. Rapidly progressive Alzheimer’s disease features distinct structures of amyloid-β. Brain 2015, 138, 1009–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simone Ruggeri, F.; Habchi, J.; Cerreta, A.; Dietler, G. AFM-Based Single Molecule Techniques: Unraveling the Amyloid Pathogenic Species. Curr. Pharm. Des. 2016, 22, 3950–3970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devitt, G.; Rice, W.; Crisford, A.; Nandhakumar, I.; Mudher, A.; Mahajan, S. Conformational Evolution of Molecular Signatures during Amyloidogenic Protein Aggregation. ACS Chem. Neurosci. 2019, 10, 4593–4611. [Google Scholar] [CrossRef]
- Festa, G.; Mallamace, F.; Sancesario, G.M.; Corsaro, C.; Mallamace, D.; Fazio, E.; Arcidiacono, L.; Garcia Sakai, V.; Senesi, R.; Preziosi, E.; et al. Aggregation States of Aβ1–40, Aβ1–42 and Aβp3–42 Amyloid Beta Peptides: A SANS Study. Int. J. Mol. Sci. 2019, 20, 4126. [Google Scholar] [CrossRef] [Green Version]
- Watanabe-Nakayama, T.; Ono, K.; Itami, M.; Takahashi, R.; Teplow, D.B.; Yamada, M. High-speed atomic force microscopy reveals structural dynamics of amyloid β 1–42 aggregates. Proc. Natl. Acad. Sci. 2016, 113, 5835–5840. [Google Scholar] [CrossRef] [Green Version]
- Gremer, L.; Schölzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril structure of amyloid-β(1–42) by cryo–electron microscopy. Science 2017, 358, 116–119. [Google Scholar] [CrossRef] [Green Version]
- Buchete, N.-V.; Tycko, R.; Hummer, G. Molecular dynamics simulations of Alzheimer’s beta-amyloid protofilaments. J. Mol. Biol. 2005, 353, 804–821. [Google Scholar] [CrossRef]
- Urbanc, B.; Cruz, L.; Ding, F.; Sammond, D.; Khare, S.; Buldyrev, S.V.; Stanley, H.E.; Dokholyan, N.V. Molecular Dynamics Simulation of Amyloid β Dimer Formation. Biophys. J. 2004, 87, 2310–2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunde, M.; Blake, C. The Structure of Amyloid Fibrils by Electron Microscopy and X-Ray Diffraction. Adv. Protein Chem. 1997, 50, 123–159. [Google Scholar] [PubMed]
- Colvin, M.T.; Silvers, R.; Ni, Q.Z.; Can, T.V.; Sergeyev, I.; Rosay, M.; Donovan, K.J.; Michael, B.; Wall, J.; Linse, S.; et al. Atomic Resolution Structure of Monomorphic Aβ 42 Amyloid Fibrils. J. Am. Chem. Soc. 2016, 138, 9663–9674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.-X.; Qiang, W.; Yau, W.-M.; Schwieters, C.D.; Meredith, S.C.; Tycko, R. Molecular Structure of β-Amyloid Fibrils in Alzheimer’s Disease Brain Tissue. Cell 2013, 154, 1257–1268. [Google Scholar] [CrossRef] [Green Version]
- Wälti, M.A.; Ravotti, F.; Arai, H.; Glabe, C.G.; Wall, J.S.; Böckmann, A.; Güntert, P.; Meier, B.H.; Riek, R. Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc. Natl. Acad. Sci. 2016, 113, E4976–E4984. [Google Scholar] [CrossRef] [Green Version]
- Luhrs, T.; Ritter, C.; Adrian, M.; Riek-Loher, D.; Bohrmann, B.; Dobeli, H.; Schubert, D.; Riek, R. 3D structure of Alzheimer’s amyloid- (1-42) fibrils. Proc. Natl. Acad. Sci. 2005, 102, 17342–17347. [Google Scholar] [CrossRef] [Green Version]
- Kollmer, M.; Close, W.; Funk, L.; Rasmussen, J.; Bsoul, A.; Schierhorn, A.; Schmidt, M.; Sigurdson, C.J.; Jucker, M.; Fändrich, M. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 2019, 10, 4760–4768. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Jin, M.; Koeglsperger, T.; Shepardson, N.E.; Shankar, G.M.; Selkoe, D.J. Soluble Aβ oligomers inhibit long-term potentiation through a mechanism involving excessive activation of extrasynaptic NR2B-containing NMDA receptors. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 6627–6638. [Google Scholar] [CrossRef]
- De Felice, F.G.; Wu, D.; Lambert, M.P.; Fernandez, S.J.; Velasco, P.T.; Lacor, P.N.; Bigio, E.H.; Jerecic, J.; Acton, P.J.; Shughrue, P.J.; et al. Alzheimer’s disease-type neuronal tau hyperphosphorylation induced by Aβ oligomers. Neurobiol. Aging 2008, 29, 1334–1347. [Google Scholar] [CrossRef] [Green Version]
- De Felice, F.G.; Velasco, P.T.; Lambert, M.P.; Viola, K.; Fernandez, S.J.; Ferreira, S.T.; Klein, W.L. Aβ Oligomers Induce Neuronal Oxidative Stress through an N -Methyl-D-aspartate Receptor-dependent Mechanism That Is Blocked by the Alzheimer Drug Memantine. J. Biol. Chem. 2007, 282, 11590–11601. [Google Scholar] [CrossRef] [Green Version]
- Tabner, B.J.; El-Agnaf, O.M.A.; Turnbull, S.; German, M.J.; Paleologou, K.E.; Hayashi, Y.; Cooper, L.J.; Fullwood, N.J.; Allsop, D. Hydrogen Peroxide Is Generated during the Very Early Stages of Aggregation of the Amyloid Peptides Implicated in Alzheimer Disease and Familial British Dementia. J. Biol. Chem. 2005, 280, 35789–35792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sponne, I.; Fifre, A.; Drouet, B.; Klein, C.; Koziel, V.; Pinçon-Raymond, M.; Olivier, J.-L.; Chambaz, J.; Pillot, T. Apoptotic Neuronal Cell Death Induced by the Non-fibrillar Amyloid-β Peptide Proceeds through an Early Reactive Oxygen Species-dependent Cytoskeleton Perturbation. J. Biol. Chem. 2003, 278, 3437–3445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, V.D.; Viola, K.L.; Klein, W.L.; Finch, C.E. Reversible Inactivation of Superoxide-Sensitive Aconitase in Aβ1-42-Treated Neuronal Cell Lines. J. Neurochem. 2002, 75, 1977–1985. [Google Scholar] [CrossRef] [PubMed]
- Tomiyama, T.; Matsuyama, S.; Iso, H.; Umeda, T.; Takuma, H.; Ohnishi, K.; Ishibashi, K.; Teraoka, R.; Sakama, N.; Yamashita, T.; et al. A Mouse Model of Amyloid Oligomers: Their Contribution to Synaptic Alteration, Abnormal Tau Phosphorylation, Glial Activation, and Neuronal Loss In Vivo. J. Neurosci. 2010, 30, 4845–4856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maezawa, I.; Zimin, P.I.; Wulff, H.; Jin, L.-W. Amyloid-β Protein Oligomer at Low Nanomolar Concentrations Activates Microglia and Induces Microglial Neurotoxicity. J. Biol. Chem. 2011, 286, 3693–3706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Akama, K.T.; Krafft, G.A.; Chromy, B.A.; Van Eldik, L.J. Amyloid-β peptide activates cultured astrocytes: Morphological alterations, cytokine induction and nitric oxide release. Brain Res. 1998, 785, 195–206. [Google Scholar] [CrossRef]
- Ferretti, M.T.; Bruno, M.A.; Ducatenzeiler, A.; Klein, W.L.; Cuello, A.C. Intracellular Aβ-oligomers and early inflammation in a model of Alzheimer’s disease. Neurobiol. Aging 2012, 33, 1329–1342. [Google Scholar] [CrossRef]
- Nishitsuji, K.; Tomiyama, T.; Ishibashi, K.; Ito, K.; Teraoka, R.; Lambert, M.P.; Klein, W.L.; Mori, H. The E693Δ Mutation in Amyloid Precursor Protein Increases Intracellular Accumulation of Amyloid β Oligomers and Causes Endoplasmic Reticulum Stress-Induced Apoptosis in Cultured Cells. Am. J. Pathol. 2009, 174, 957–969. [Google Scholar] [CrossRef] [Green Version]
- Umeda, T.; Tomiyama, T.; Sakama, N.; Tanaka, S.; Lambert, M.P.; Klein, W.L.; Mori, H. Intraneuronal amyloid β oligomers cause cell death via endoplasmic reticulum stress, endosomal/lysosomal leakage, and mitochondrial dysfunction in vivo. J. Neurosci. Res. 2011, 89, 1031–1042. [Google Scholar] [CrossRef]
- Resende, R.; Ferreiro, E.; Pereira, C.; Oliveira, C.R. ER stress is involved in Aβ--induced GSK--3β activation and tau phosphorylation. J. Neurosci. Res. 2008, 86, 2091–2099. [Google Scholar] [CrossRef] [Green Version]
- Shankar, G.M.; Bloodgood, B.L.; Townsend, M.; Walsh, D.M.; Selkoe, D.J.; Sabatini, B.L. Natural Oligomers of the Alzheimer Amyloid- Protein Induce Reversible Synapse Loss by Modulating an NMDA-Type Glutamate Receptor-Dependent Signaling Pathway. J. Neurosci. 2007, 27, 2866–2875. [Google Scholar] [CrossRef] [PubMed]
- Lacor, P.N.; Buniel, M.C.; Furlow, P.W.; Sanz Clemente, A.; Velasco, P.T.; Wood, M.; Viola, K.L.; Klein, W.L. A Oligomer-Induced Aberrations in Synapse Composition, Shape, and Density Provide a Molecular Basis for Loss of Connectivity in Alzheimer’s Disease. J. Neurosci. 2007, 27, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Pickett, E.K.; Koffie, R.M.; Wegmann, S.; Henstridge, C.M.; Herrmann, A.G.; Colom-Cadena, M.; Lleo, A.; Kay, K.R.; Vaught, M.; Soberman, R.; et al. Non-Fibrillar Oligomeric Amyloid-β within Synapses. J. Alzheimers Dis. 2016, 53, 787–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomiyama, T.; Nagata, T.; Shimada, H.; Teraoka, R.; Fukushima, A.; Kanemitsu, H.; Takuma, H.; Kuwano, R.; Imagawa, M.; Ataka, S.; et al. A new amyloid β variant favoring oligomerization in Alzheimer’s-type dementia. Ann. Neurol. 2008, 63, 377–387. [Google Scholar] [CrossRef]
- Shimada, H.; Ataka, S.; Tomiyama, T.; Takechi, H.; Mori, H.; Miki, T. Clinical Course of Patients with Familial Early-Onset Alzheimer’s Disease Potentially Lacking Senile Plaques Bearing the E693Δ Mutation in Amyloid Precursor Protein. Dement. Geriatr. Cogn. Disord. 2011, 32, 45–54. [Google Scholar] [CrossRef]
- Inayathullah, M.; Teplow, D.B. Structural dynamics of the ΔE22 (Osaka) familial Alzheimer’s disease-linked amyloid β-protein. Amyloid 2011, 18, 98–107. [Google Scholar] [CrossRef]
- Lambert, M.P.; Velasco, P.T.; Chang, L.; Viola, K.L.; Fernandez, S.; Lacor, P.N.; Khuon, D.; Gong, Y.; Bigio, E.H.; Shaw, P.; et al. Monoclonal antibodies that target pathological assemblies of Aβ. J. Neurochem. 2007, 100, 23–35. [Google Scholar] [CrossRef]
- Zameer, A.; Kasturirangan, S.; Emadi, S.; Nimmagadda, S.V.; Sierks, M.R. Anti-oligomeric Aβ Single-chain Variable Domain Antibody Blocks Aβ-induced Toxicity Against Human Neuroblastoma Cells. J. Mol. Biol. 2008, 384, 917–928. [Google Scholar] [CrossRef]
- Klyubin, I.; Walsh, D.M.; Lemere, C.A.; Cullen, W.K.; Shankar, G.M.; Betts, V.; Spooner, E.T.; Jiang, L.; Anwyl, R.; Selkoe, D.J.; et al. Amyloid β protein immunotherapy neutralizes Aβ oligomers that disrupt synaptic plasticity in vivo. Nat. Med. 2005, 11, 556–561. [Google Scholar] [CrossRef]
- Xiao, C.; Davis, F.J.; Chauhan, B.C.; Viola, K.L.; Lacor, P.N.; Velasco, P.T.; Klein, W.L.; Chauhan, N.B. Brain Transit and Ameliorative Effects of Intranasally Delivered Anti-Amyloid-β Oligomer Antibody in 5XFAD Mice. J. Alzheimers Dis. 2013, 35, 777–788. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Wang, S.; Wang, Y.; Zhang, R.; Li, Y.; Su, Y.; Zhou, W.; Yu, X.; Liu, R. Pan-Amyloid Oligomer Specific scFv Antibody Attenuates Memory Deficits and Brain Amyloid Burden in Mice with Alzheimer’s Disease. Curr. Alzheimer Res. 2014, 11, 69–78. [Google Scholar] [CrossRef]
- Sebollela, A.; Cline, E.N.; Popova, I.; Luo, K.; Sun, X.; Ahn, J.; Barcelos, M.A.; Bezerra, V.N.; Lyra e Silva, N.M.; Patel, J.; et al. A human scFv antibody that targets and neutralizes high molecular weight pathogenic amyloid-β oligomers. J. Neurochem. 2017, 142, 934–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arispe, N. Architecture of the Alzheimer’s A beta P Ion Channel Pore. J. Membr. Biol. 2004, 197, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Evangelisti, E.; Wright, D.; Zampagni, M.; Cascella, R.; Fiorillo, C.; Bagnoli, S.; Relini, A.; Nichino, D.; Scartabelli, T.; Nacmias, B.; et al. Lipid Rafts Mediate Amyloid-Induced Calcium Dyshomeostasis and Oxidative Stress in Alzheimer’s Disease. Curr. Alzheimer Res. 2013, 10, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacor, P.N. Synaptic Targeting by Alzheimer’s-Related Amyloid Oligomers. J. Neurosci. 2004, 24, 10191–10200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, Y.; Chang, L.; Viola, K.L.; Lacor, P.N.; Lambert, M.P.; Finch, C.E.; Krafft, G.A.; Klein, W.L. Alzheimer’s disease-affected brain: Presence of oligomeric A ligands (ADDLs) suggests a molecular basis for reversible memory loss. Proc. Natl. Acad. Sci. 2003, 100, 10417–10422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.-Q.; De Felice, F.G.; Fernandez, S.; Chen, H.; Lambert, M.P.; Quon, M.J.; Krafft, G.A.; Klein, W.L. Amyloid beta oligomers induce impairment of neuronal insulin receptors. FASEB J. 2008, 22, 246–260. [Google Scholar] [CrossRef] [Green Version]
- Renner, M.; Lacor, P.N.; Velasco, P.T.; Xu, J.; Contractor, A.; Klein, W.L.; Triller, A. Deleterious Effects of Amyloid β Oligomers Acting as an Extracellular Scaffold for mGluR5. Neuron 2010, 66, 739–754. [Google Scholar] [CrossRef] [Green Version]
- Thathiah, A.; De Strooper, B. The role of G protein-coupled receptors in the pathology of Alzheimer’s disease. Nat. Rev. Neurosci. 2011, 12, 73–87. [Google Scholar] [CrossRef]
- Godoy, J.A.; Rios, J.A.; Zolezzi, J.M.; Braidy, N.; Inestrosa, N.C. Signaling pathway cross talk in Alzheimer’s disease. Cell Commun. Signal. 2014, 12, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Magdesian, M.H.; Carvalho, M.M.V.F.; Mendes, F.A.; Saraiva, L.M.; Juliano, M.A.; Juliano, L.; Garcia-Abreu, J.; Ferreira, S.T. Amyloid-β Binds to the Extracellular Cysteine-rich Domain of Frizzled and Inhibits Wnt/β-Catenin Signaling. J. Biol. Chem. 2008, 283, 9359–9368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Zhao, J.; Zhang, X.; Wang, S.; Viola, K.L.; Chow, F.E.; Zhang, Y.; Lippa, C.; Klein, W.L.; Gong, Y. Amyloid Beta Oligomers Target to Extracellular and Intracellular Neuronal Synaptic Proteins in Alzheimer’s Disease. Front. Neurol. 2019, 10, 1140–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, L.M.; Kostylev, M.A.; Lee, S.; Strittmatter, S.M. Systematic and standardized comparison of reported amyloid-β receptors for sufficiency, affinity, and Alzheimer’s disease relevance. J. Biol. Chem. 2019, 294, 6042–6053. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, H.B.; van Dyck, C.H.; Strittmatter, S.M. Fyn kinase inhibition as a novel therapy for Alzheimer’s disease. Alzheimers Res. Ther. 2014, 6, 8–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhao, Y.; Zhang, L.; Yu, W.; Wang, Y.; Chang, W. Cellular Prion Protein as a Receptor of Toxic Amyloid-β42 Oligomers Is Important for Alzheimer’s Disease. Front. Cell. Neurosci. 2019, 13, 339–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar, S.V.; Gallardo, C.; Kaufman, A.C.; Herber, C.S.; Haas, L.T.; Robinson, S.; Manson, J.C.; Lee, M.K.; Strittmatter, S.M. Conditional Deletion of Prnp Rescues Behavioral and Synaptic Deficits after Disease Onset in Transgenic Alzheimer’s Disease. J. Neurosci. 2017, 37, 9207–9221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohnishi, T.; Yanazawa, M.; Sasahara, T.; Kitamura, Y.; Hiroaki, H.; Fukazawa, Y.; Kii, I.; Nishiyama, T.; Kakita, A.; Takeda, H.; et al. Na, K-ATPase α3 is a death target of Alzheimer patient amyloid-β assembly. Proc. Natl. Acad. Sci. 2015, 112, E4465–E4474. [Google Scholar] [CrossRef] [Green Version]
- DiChiara, T.; DiNunno, N.; Clark, J.; Bu, R.L.; Cline, E.N.; Rollins, M.G.; Gong, Y.; Brody, D.L.; Sligar, S.G.; Velasco, P.T.; et al. Alzheimer’s Toxic Amyloid Beta Oligomers: Unwelcome Visitors to the Na/K ATPase alpha3 Docking Station. Yale J. Biol. Med. 2017, 90, 45–61. [Google Scholar]
- Zhang, F.; Gannon, M.; Chen, Y.; Yan, S.; Zhang, S.; Feng, W.; Tao, J.; Sha, B.; Liu, Z.; Saito, T.; et al. β-amyloid redirects norepinephrine signaling to activate the pathogenic GSK3β/tau cascade. Sci. Transl. Med. 2020, 12, eaay6931. [Google Scholar] [CrossRef]
- Findley, C.A.; Bartke, A.; Hascup, K.N.; Hascup, E.R. Amyloid Beta-Related Alterations to Glutamate Signaling Dynamics During Alzheimer’s Disease Progression. ASN Neuro 2019, 11. [Google Scholar] [CrossRef]
- Goulay, R.; Mena Romo, L.; Hol, E.M.; Dijkhuizen, R.M. From Stroke to Dementia: A Comprehensive Review Exposing Tight Interactions Between Stroke and Amyloid-β Formation. Transl. Stroke Res. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wu, X.; Li, X.; Jiang, L.-L.; Gui, X.; Liu, Y.; Sun, Y.; Zhu, B.; Piña-Crespo, J.C.; Zhang, M.; et al. TREM2 Is a Receptor for β-Amyloid that Mediates Microglial Function. Neuron 2018, 97, 1023–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, P.; Shah, J. Role of Vitamin D in Amyloid clearance via LRP-1 upregulation in Alzheimer’s disease: A potential therapeutic target? J. Chem. Neuroanat. 2017, 85, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Bückig, A.; Tikkanen, R.; Herzog, V.; Schmitz, A. Cytosolic and nuclear aggregation of the amyloid β-peptide following its expression in the endoplasmic reticulum. Histochem. Cell Biol. 2002, 118, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Gouras, G.K.; Tsai, J.; Naslund, J.; Vincent, B.; Edgar, M.; Checler, F.; Greenfield, J.P.; Haroutunian, V.; Buxbaum, J.D.; Xu, H.; et al. Intraneuronal Aβ42 Accumulation in Human Brain. Am. J. Pathol. 2000, 156, 15–20. [Google Scholar] [CrossRef]
- Bayer, T.A.; Wirths, O. Intracellular accumulation of amyloid-beta – a predictor for synaptic dysfunction and neuron loss in Alzheimer’s disease. Front. Aging Neurosci. 2010, 2, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.H.; Nagao, T.; Gouras, G.K. Plaque formation and the intraneuronal accumulation of β-amyloid in Alzheimer’s disease: Intraneuronal accumulation of β-amyloid. Pathol. Int. 2017, 67, 185–193. [Google Scholar] [CrossRef]
- Brewer, G.J.; Herrera, R.A.; Philipp, S.; Sosna, J.; Reyes-Ruiz, J.M.; Glabe, C.G. Age-Related Intraneuronal Aggregation of Amyloid-β in Endosomes, Mitochondria, Autophagosomes, and Lysosomes. J. Alzheimers Dis. 2020, 73, 229–246. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, T.; Bieger, S.C.; Brühl, B.; Tienari, P.J.; Ida, N.; Allsop, D.; Roberts, G.W.; Masters, C.L.; Dotti, C.G.; Unsicker, K.; et al. Distinct sites of intracellular production for Alzheimer’s disease Aβ40/42 amyloid peptides. Nat. Med. 1997, 3, 1016–1020. [Google Scholar] [CrossRef]
- Pacheco-Quinto, J.; Eckman, E.A. Endothelin-converting Enzymes Degrade Intracellular β-Amyloid Produced within the Endosomal/Lysosomal Pathway and Autophagosomes. J. Biol. Chem. 2013, 288, 5606–5615. [Google Scholar] [CrossRef] [Green Version]
- Lacosta, A.-M.; Insua, D.; Badi, H.; Pesini, P.; Sarasa, M. Neurofibrillary Tangles of Aβx-40 in Alzheimer’s Disease Brains. J. Alzheimers Dis. 2017, 58, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-L.; Wang, W.-A.; Tan, J.-X.; Huang, J.-K.; Zhang, X.; Zhang, B.-Z.; Wang, Y.-H.; YangCheng, H.-Y.; Zhu, H.-L.; Sun, X.-J.; et al. Expression of -Amyloid Induced Age-Dependent Presynaptic and Axonal Changes in Drosophila. J. Neurosci. 2010, 30, 1512–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsson, T.T.; Klementieva, O.; Gouras, G.K. Prion-like seeding and nucleation of intracellular amyloid-β. Neurobiol. Dis. 2018, 113, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Luehmann, M.; Spires-Jones, T.L.; Prada, C.; Garcia-Alloza, M.; de Calignon, A.; Rozkalne, A.; Koenigsknecht-Talboo, J.; Holtzman, D.M.; Bacskai, B.J.; Hyman, B.T. Rapid appearance and local toxicity of amyloid-β plaques in a mouse model of Alzheimer’s disease. Nature 2008, 451, 720–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Querol-Vilaseca, M.; Colom-Cadena, M.; Pegueroles, J.; Nuñez-Llaves, R.; Luque-Cabecerans, J.; Muñoz-Llahuna, L.; Andilla, J.; Belbin, O.; Spires-Jones, T.L.; Gelpi, E.; et al. Nanoscale structure of amyloid-β plaques in Alzheimer’s disease. Sci. Rep. 2019, 9, 5181–5191. [Google Scholar] [CrossRef] [Green Version]
- Masters, C.L.; Multhaup, G.; Simms, G.; Pottgiesser, J.; Martins, R.N.; Beyreuther, K. Neuronal origin of a cerebral amyloid: Neurofibrillary tangles of Alzheimer’s disease contain the same protein as the amyloid of plaque cores and blood vessels. EMBO J. 1985, 4, 2757–2763. [Google Scholar] [CrossRef]
- Gkanatsiou, E.; Portelius, E.; Toomey, C.E.; Blennow, K.; Zetterberg, H.; Lashley, T.; Brinkmalm, G. A distinct brain beta amyloid signature in cerebral amyloid angiopathy compared to Alzheimer’s disease. Neurosci. Lett. 2019, 701, 125–131. [Google Scholar] [CrossRef]
- Tapia-Rojas, C.; Cabezas-Opazo, F.; Deaton, C.A.; Vergara, E.H.; Johnson, G.V.W.; Quintanilla, R.A. It’s all about tau. Prog. Neurobiol. 2019, 175, 54–76. [Google Scholar] [CrossRef]
- Chong, F.P.; Ng, K.Y.; Koh, R.Y.; Chye, S.M. Tau Proteins and Tauopathies in Alzheimer’s Disease. Cell. Mol. Neurobiol. 2018, 38, 965–980. [Google Scholar] [CrossRef]
- Mroczko; Groblewska; Litman-Zawadzka The Role of Protein Misfolding and Tau Oligomers (TauOs) in Alzheimer′s Disease (AD). Int. J. Mol. Sci. 2019, 20, 4661. [CrossRef] [Green Version]
- Almansoub, H.A.M.M.; Tang, H.; Wu, Y.; Wang, D.-Q.; Mahaman, Y.A.R.; Wei, N.; Almansob, Y.A.M.; He, W.; Liu, D. Tau Abnormalities and the Potential Therapy in Alzheimer’s Disease. J. Alzheimers Dis. 2019, 67, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Arakhamia, T.; Lee, C.E.; Carlomagno, Y.; Duong, D.M.; Kundinger, S.R.; Wang, K.; Williams, D.; DeTure, M.; Dickson, D.W.; Cook, C.N.; et al. Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell 2020, 180, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, N.; Bretteville, A.; Hamdane, M.; Caillet-Boudin, M.-L.; Grognet, P.; Bombois, S.; Blum, D.; Delacourte, A.; Pasquier, F.; Vanmechelen, E.; et al. Biochemistry of Tau in Alzheimer’s disease and related neurological disorders. Expert Rev. Proteomics 2008, 5, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Kabat, J.; Novak, M.; Wu, Q.; Grundke-Iqbal, I.; Iqbal, K. Phosphorylation of Tau at Both Thr 231 and Ser 262 Is Required for Maximal Inhibition of Its Binding to Microtubules. Arch. Biochem. Biophys. 1998, 357, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Dickey, C.A.; Kamal, A.; Lundgren, K.; Klosak, N.; Bailey, R.M.; Dunmore, J.; Ash, P.; Shoraka, S.; Zlatkovic, J.; Eckman, C.B.; et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 2007, 117, 648–658. [Google Scholar] [CrossRef] [Green Version]
- Barthélemy, N.R.; Li, Y.; Joseph-Mathurin, N.; Gordon, B.A.; Hassenstab, J.; Benzinger, T.L.S.; Buckles, V.; Fagan, A.M.; Perrin, R.J.; Goate, A.M.; et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat. Med. 2020, 26, 398–407. [Google Scholar] [CrossRef]
- Dawson, H.N.; Ferreira, A.; Eyster, M.V.; Ghoshal, N.; Binder, L.I.; Vitek, M.P. Inhibition of neuronal maturation in primary hippocampal neurons from tau deficient mice. J. Cell Sci. 2001, 114, 1179–1187. [Google Scholar]
- Lei, P.; Ayton, S.; Finkelstein, D.I.; Spoerri, L.; Ciccotosto, G.D.; Wright, D.K.; Wong, B.X.W.; Adlard, P.A.; Cherny, R.A.; Lam, L.Q.; et al. Tau deficiency induces parkinsonism with dementia by impairing APP-mediated iron export. Nat. Med. 2012, 18, 291–295. [Google Scholar] [CrossRef]
- Ma, Q.-L.; Zuo, X.; Yang, F.; Ubeda, O.J.; Gant, D.J.; Alaverdyan, M.; Kiosea, N.C.; Nazari, S.; Chen, P.P.; Nothias, F.; et al. Loss of MAP Function Leads to Hippocampal Synapse Loss and Deficits in the Morris Water Maze with Aging. J. Neurosci. 2014, 34, 7124–7136. [Google Scholar] [CrossRef] [Green Version]
- Bi, M.; Gladbach, A.; van Eersel, J.; Ittner, A.; Przybyla, M.; van Hummel, A.; Chua, S.W.; van der Hoven, J.; Lee, W.S.; Müller, J.; et al. Tau exacerbates excitotoxic brain damage in an animal model of stroke. Nat. Commun. 2017, 8, 473–488. [Google Scholar] [CrossRef] [Green Version]
- Tuo, Q.; Lei, P.; Jackman, K.A.; Li, X.; Xiong, H.; Li, X.; Liuyang, Z.; Roisman, L.; Zhang, S.; Ayton, S.; et al. Tau-mediated iron export prevents ferroptotic damage after ischemic stroke. Mol. Psychiatry 2017, 22, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Naseri, N.N.; Wang, H.; Guo, J.; Sharma, M.; Luo, W. The complexity of tau in Alzheimer’s disease. Neurosci. Lett. 2019, 705, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Sultan, A.; Nesslany, F.; Violet, M.; Bégard, S.; Loyens, A.; Talahari, S.; Mansuroglu, Z.; Marzin, D.; Sergeant, N.; Humez, S.; et al. Nuclear Tau, a Key Player in Neuronal DNA Protection. J. Biol. Chem. 2011, 286, 4566–4575. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.; Newman, S.T.; Gard, D.L.; Band, H.; Panchamoorthy, G. Tau interacts with src-family non-receptor tyrosine kinases. J. Cell Sci. 1998, 111, 3167–3177. [Google Scholar]
- Ittner, L.M.; Ke, Y.D.; Delerue, F.; Bi, M.; Gladbach, A.; van Eersel, J.; Wölfing, H.; Chieng, B.C.; Christie, M.J.; Napier, I.A.; et al. Dendritic Function of Tau Mediates Amyloid-β Toxicity in Alzheimer’s Disease Mouse Models. Cell 2010, 142, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pooler, A.M.; Noble, W.; Hanger, D.P. A role for tau at the synapse in Alzheimer’s disease pathogenesis. Neuropharmacology 2014, 76, 1–8. [Google Scholar] [CrossRef]
- Gong, C.-X.; Liu, F.; Grundke-Iqbal, I.; Iqbal, K. Post-translational modifications of tau protein in Alzheimer’s disease. J. Neural Transm. 2005, 112, 813–838. [Google Scholar] [CrossRef]
- Marcus, J.N.; Schachter, J. Targeting Post-translational Modifications on Tau as a Therapeutic Strategy for Alzheimer’s Disease. J. Neurogenet. 2011, 25, 127–133. [Google Scholar] [CrossRef]
- Hervy, J.; Bicout, D.J. Dynamical decoration of stabilized-microtubules by Tau-proteins. Sci. Rep. 2019, 9, 12473–12488. [Google Scholar] [CrossRef] [Green Version]
- Yeboah, F.; Kim, T.-E.; Bill, A.; Dettmer, U. Dynamic behaviors of α-synuclein and tau in the cellular context: New mechanistic insights and therapeutic opportunities in neurodegeneration. Neurobiol. Dis. 2019, 132, 104543–104558. [Google Scholar] [CrossRef]
- Kolarova, M.; García-Sierra, F.; Bartos, A.; Ricny, J.; Ripova, D. Structure and Pathology of Tau Protein in Alzheimer Disease. Int. J. Alzheimers Dis. 2012, 2012, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandelkow, E.-M.; Biernat, J.; Drewes, G.; Gustke, N.; Trinczek, B.; Mandelkow, E. Tau domains, phosphorylation, and interactions with microtubules. Neurobiol. Aging 1995, 16, 355–362. [Google Scholar] [CrossRef]
- Brady, S.T.; Sperry, A.O. Biochemical and functional diversity of microtubule motors in the nervous system. Curr. Opin. Neurobiol. 1995, 5, 551–558. [Google Scholar] [CrossRef]
- Baas, P.W.; Qiang, L. Tau: It’s Not What You Think. Trends Cell Biol. 2019, 29, 452–461. [Google Scholar] [CrossRef]
- Mandelkow, E.-M.; Mandelkow, E. Biochemistry and Cell Biology of Tau Protein in Neurofibrillary Degeneration. Cold Spring Harb. Perspect. Med. 2012, 2, a006247–a006273. [Google Scholar] [CrossRef]
- Kellogg, E.H.; Hejab, N.M.A.; Poepsel, S.; Downing, K.H.; DiMaio, F.; Nogales, E. Near-atomic model of microtubule-tau interactions. Science 2018, 360, 1242–1246. [Google Scholar] [CrossRef] [Green Version]
- Dixit, R.; Ross, J.L.; Goldman, Y.E.; Holzbaur, E.L.F. Differential Regulation of Dynein and Kinesin Motor Proteins by Tau. Science 2008, 319, 1086–1089. [Google Scholar] [CrossRef] [Green Version]
- Reddy, P.H. Abnormal tau, mitochondrial dysfunction, impaired axonal transport of mitochondria, and synaptic deprivation in Alzheimer’s disease. Brain Res. 2011, 1415, 136–148. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Zhou, R.; Wu, Z.; Carrasco, M.A.; Kurshan, P.T.; Farley, J.E.; Simon, D.J.; Wang, G.; Han, B.; Hao, J.; et al. Prevalent presence of periodic actin–spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc. Natl. Acad. Sci. 2016, 113, 6029–6034. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Zhang, C.; Zhao, Q.; Li, D. Spectrin: Structure, function and disease. Sci. China Life Sci. 2013, 56, 1076–1085. [Google Scholar] [CrossRef] [Green Version]
- Morris, M.; Maeda, S.; Vossel, K.; Mucke, L. The Many Faces of Tau. Neuron 2011, 70, 410–426. [Google Scholar] [CrossRef] [Green Version]
- Marciniak, E.; Leboucher, A.; Caron, E.; Ahmed, T.; Tailleux, A.; Dumont, J.; Issad, T.; Gerhardt, E.; Pagesy, P.; Vileno, M.; et al. Tau deletion promotes brain insulin resistance. J. Exp. Med. 2017, 214, 2257–2269. [Google Scholar] [CrossRef] [PubMed]
- Shafiei, S.S.; Guerrero-Muñoz, M.J.; Castillo-Carranza, D.L. Tau Oligomers: Cytotoxicity, Propagation, and Mitochondrial Damage. Front. Aging Neurosci. 2017, 9, 83–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frandemiche, M.L.; De Seranno, S.; Rush, T.; Borel, E.; Elie, A.; Arnal, I.; Lante, F.; Buisson, A. Activity-Dependent Tau Protein Translocation to Excitatory Synapse Is Disrupted by Exposure to Amyloid-Beta Oligomers. J. Neurosci. 2014, 34, 6084–6097. [Google Scholar] [CrossRef] [PubMed]
- Friedhoff, P.; von Bergen, M.; Mandelkow, E.-M.; Mandelkow, E. Structure of tau protein and assembly into paired helical filaments. Biochim. Biophys. BBA - Mol. Basis Dis. 2000, 1502, 122–132. [Google Scholar] [CrossRef]
- Mukrasch, M.D.; Bibow, S.; Korukottu, J.; Jeganathan, S.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Structural Polymorphism of 441-Residue Tau at Single Residue Resolution. PLoS Biol. 2009, 7, e1000034. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S. Progression of Alzheimer’s disease, tau propagation, and its modifiable risk factors. Neurosci. Res. 2019, 141, 36–42. [Google Scholar] [CrossRef]
- Fanni, A.M.; Vander Zanden, C.M.; Majewska, P.V.; Majewski, J.; Chi, E.Y. Membrane-mediated fibrillation and toxicity of the tau hexapeptide PHF6. J. Biol. Chem. 2019, 294, 15304–15317. [Google Scholar] [CrossRef]
- Maeda, S.; Sahara, N.; Saito, Y.; Murayama, S.; Ikai, A.; Takashima, A. Increased levels of granular tau oligomers: An early sign of brain aging and Alzheimer’s disease. Neurosci. Res. 2006, 54, 197–201. [Google Scholar] [CrossRef]
- Griner, S.L.; Seidler, P.; Bowler, J.; Murray, K.A.; Yang, T.P.; Sahay, S.; Sawaya, M.R.; Cascio, D.; Rodriguez, J.A.; Philipp, S.; et al. Structure-based inhibitors of amyloid beta core suggest a common interface with tau. eLife 2019, 8, e46924–e46952. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zivanov, J.; Zhang, W.; Murzin, A.G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Newell, K.L.; Ghetti, B.; Goedert, M.; et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 2019, 568, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Tau filaments in neurodegenerative diseases. FEBS Lett. 2018, 592, 2383–2391. [Google Scholar] [CrossRef]
- La Joie, R.; Visani, A.V.; Baker, S.L.; Brown, J.A.; Bourakova, V.; Cha, J.; Chaudhary, K.; Edwards, L.; Iaccarino, L.; Janabi, M.; et al. Prospective longitudinal atrophy in Alzheimer’s disease correlates with the intensity and topography of baseline tau-PET. Sci. Transl. Med. 2020, 12, 5732–5745. [Google Scholar] [CrossRef]
- Guo, W.; Stoklund Dittlau, K.; Van Den Bosch, L. Axonal transport defects and neurodegeneration: Molecular mechanisms and therapeutic implications. Semin. Cell Dev. Biol. 2019, 99, 133–150. [Google Scholar] [CrossRef]
- Violet, M.; Chauderlier, A.; Delattre, L.; Tardivel, M.; Chouala, M.S.; Sultan, A.; Marciniak, E.; Humez, S.; Binder, L.; Kayed, R.; et al. Prefibrillar Tau oligomers alter the nucleic acid protective function of Tau in hippocampal neurons in vivo. Neurobiol. Dis. 2015, 82, 540–551. [Google Scholar] [CrossRef]
- Eftekharzadeh, B.; Daigle, J.G.; Kapinos, L.E.; Coyne, A.; Schiantarelli, J.; Carlomagno, Y.; Cook, C.; Miller, S.J.; Dujardin, S.; Amaral, A.S.; et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 2018, 99, 925–940. [Google Scholar] [CrossRef] [Green Version]
- Chin, J.; Scharfman, H.E. Shared cognitive and behavioral impairments in epilepsy and Alzheimer’s disease and potential underlying mechanisms. Epilepsy Behav. 2013, 26, 343–351. [Google Scholar] [CrossRef] [Green Version]
- DeVos, S.L.; Goncharoff, D.K.; Chen, G.; Kebodeaux, C.S.; Yamada, K.; Stewart, F.R.; Schuler, D.R.; Maloney, S.E.; Wozniak, D.F.; Rigo, F.; et al. Antisense Reduction of Tau in Adult Mice Protects against Seizures. J. Neurosci. 2013, 33, 12887–12897. [Google Scholar] [CrossRef] [Green Version]
- Gheyara, A.L.; Ponnusamy, R.; Djukic, B.; Craft, R.J.; Ho, K.; Guo, W.; Finucane, M.M.; Sanchez, P.E.; Mucke, L. Tau reduction prevents disease in a mouse model of Dravet syndrome: Tau Ablation in DS. Ann. Neurol. 2014, 76, 443–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tai, H.-C.; Wang, B.Y.; Serrano-Pozo, A.; Frosch, M.P.; Spires-Jones, T.L.; Hyman, B.T. Frequent and symmetric deposition of misfolded tau oligomers within presynaptic and postsynaptic terminals in Alzheimer’s disease. Acta Neuropathol. Commun. 2014, 2, 146–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zempel, H.; Dennissen, F.J.A.; Kumar, Y.; Luedtke, J.; Biernat, J.; Mandelkow, E.-M.; Mandelkow, E. Axodendritic sorting and pathological missorting of Tau are isoform-specific and determined by axon initial segment architecture. J. Biol. Chem. 2017, 292, 12192–12207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busche, M.A.; Wegmann, S.; Dujardin, S.; Commins, C.; Schiantarelli, J.; Klickstein, N.; Kamath, T.V.; Carlson, G.A.; Nelken, I.; Hyman, B.T. Tau impairs neural circuits, dominating amyloid-β effects, in Alzheimer models in vivo. Nat. Neurosci. 2019, 22, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Terada, T.; Yokokura, M.; Obi, T.; Bunai, T.; Yoshikawa, E.; Ando, I.; Shimada, H.; Suhara, T.; Higuchi, M.; Ouchi, Y. In vivo direct relation of tau pathology with neuroinflammation in early Alzheimer’s disease. J. Neurol. 2019, 266, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Vogels, T.; Murgoci, A.-N.; Hromádka, T. Intersection of pathological tau and microglia at the synapse. Acta Neuropathol. Commun. 2019, 7, 109–134. [Google Scholar] [CrossRef]
- Pérez, M.; Avila, J.; Hernández, F. Propagation of Tau via Extracellular Vesicles. Front. Neurosci. 2019, 13, 698–705. [Google Scholar] [CrossRef]
- Dujardin, S.; Bégard, S.; Caillierez, R.; Lachaud, C.; Delattre, L.; Carrier, S.; Loyens, A.; Galas, M.-C.; Bousset, L.; Melki, R.; et al. Ectosomes: A New Mechanism for Non-Exosomal Secretion of Tau Protein. PLoS ONE 2014, 9, e100760. [Google Scholar] [CrossRef]
- Vasili, E.; Dominguez-Meijide, A.; Outeiro, T.F. Spreading of α-Synuclein and Tau: A Systematic Comparison of the Mechanisms Involved. Front. Mol. Neurosci. 2019, 12, 107–130. [Google Scholar] [CrossRef] [Green Version]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Sengupta, U.; Guerrero-Munoz, M.J.; Kiritoshi, T.; Neugebauer, V.; Jackson, G.R.; Kayed, R. Alzheimer brain-derived tau oligomers propagate pathology from endogenous tau. Sci. Rep. 2012, 2, 700–707. [Google Scholar] [CrossRef]
- Gibbons, G.S.; Lee, V.M.Y.; Trojanowski, J.Q. Mechanisms of Cell-to-Cell Transmission of Pathological Tau: A Review. JAMA Neurol. 2019, 76, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Franzmeier, N.; Rubinski, A.; Neitzel, J.; Kim, Y.; Damm, A.; Na, D.L.; Kim, H.J.; Lyoo, C.H.; Cho, H.; Finsterwalder, S.; et al. Functional connectivity associated with tau levels in ageing, Alzheimer’s, and small vessel disease. Brain 2019, 142, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J. The discovery of Alzheimer--causing mutations in the APP gene and the formulation of the “amyloid cascade hypothesis. ” FEBS J. 2017, 284, 1040–1044. [Google Scholar] [CrossRef]
- Di Fede, G.; Catania, M.; Morbin, M.; Rossi, G.; Suardi, S.; Mazzoleni, G.; Merlin, M.; Giovagnoli, A.R.; Prioni, S.; Erbetta, A.; et al. A Recessive Mutation in the APP Gene with Dominant-Negative Effect on Amyloidogenesis. Science 2009, 323, 1473–1477. [Google Scholar] [CrossRef] [Green Version]
- Jonsson, T.; Atwal, J.K.; Steinberg, S.; Snaedal, J.; Jonsson, P.V.; Bjornsson, S.; Stefansson, H.; Sulem, P.; Gudbjartsson, D.; Maloney, J.; et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 2012, 488, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Condron, M.M.; Teplow, D.B. Effects of the English (H6R) and Tottori (D7N) Familial Alzheimer Disease Mutations on Amyloid β-Protein Assembly and Toxicity. J. Biol. Chem. 2010, 285, 23186–23197. [Google Scholar] [CrossRef] [Green Version]
- Obici, L.; Demarchi, A.; de Rosa, G.; Bellotti, V.; Marciano, S.; Donadei, S.; Arbustini, E.; Palladini, G.; Diegoli, M.; Genovese, E.; et al. A novelAβPP mutation exclusively associated with cerebral amyloid angiopathy. Ann. Neurol. 2005, 58, 639–644. [Google Scholar] [CrossRef]
- Stocker, H.; Möllers, T.; Perna, L.; Brenner, H. The genetic risk of Alzheimer’s disease beyond APOE ε4: Systematic review of Alzheimer’s genetic risk scores. Transl. Psychiatry 2018, 8, 166–175. [Google Scholar] [CrossRef]
- Huynh, T.-P.V.; Davis, A.A.; Ulrich, J.D.; Holtzman, D.M. Apolipoprotein E and Alzheimer’s disease: The influence of apolipoprotein E on amyloid-β and other amyloidogenic proteins. J. Lipid Res. 2017, 58, 824–836. [Google Scholar] [CrossRef] [Green Version]
- Holtzman, D.M.; Bales, K.R.; Tenkova, T.; Fagan, A.M.; Parsadanian, M.; Sartorius, L.J.; Mackey, B.; Olney, J.; McKeel, D.; Wozniak, D.; et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. 2000, 97, 2892–2897. [Google Scholar] [CrossRef] [Green Version]
- de Rojas, I.; Moreno-Grau, S.; Tesi, N.; Grenier-Boley, B.; Andrade, V.; Jansen, I.; Pedersen, N.L.; Stringa, N.; Zettergren, A.; Hernández, I.; et al. Common variants in Alzheimer’s disease: Novel association of six genetic variants with AD and risk stratification by polygenic risk scores. Mol. Genet. Genom. Med. 2019. [Google Scholar] [CrossRef] [Green Version]
- Sierksma, A.; Lu, A.; Mancuso, R.; Fattorelli, N.; Thrupp, N.; Salta, E.; Zoco, J.; Blum, D.; Buée, L.; De Strooper, B.; et al. Novel Alzheimer risk genes determine the microglia response to amyloid--β but not to TAU pathology. EMBO Mol. Med. 2020, 12, e10606–e10624. [Google Scholar] [CrossRef] [PubMed]
- van der Kant, R.; Goldstein, L.S.B.; Ossenkoppele, R. Amyloid-β-independent regulators of tau pathology in Alzheimer disease. Nat. Rev. Neurosci. 2020, 21, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.-H.; Bastianetto, S.; Mennicken, F.; Ma, W.; Kar, S. Amyloid β peptide induces tau phosphorylation and loss of cholinergic neurons in rat primary septal cultures. Neuroscience 2002, 115, 201–211. [Google Scholar] [CrossRef]
- Jin, M.; O’Nuallain, B.; Hong, W.; Boyd, J.; Lagomarsino, V.N.; O’Malley, T.T.; Liu, W.; Vanderburg, C.R.; Frosch, M.P.; Young-Pearse, T.; et al. An in vitro paradigm to assess potential anti-Aβ antibodies for Alzheimer’s disease. Nat. Commun. 2018, 9, 2676–2689. [Google Scholar] [CrossRef]
- Sato, C.; Barthélemy, N.R.; Mawuenyega, K.G.; Patterson, B.W.; Gordon, B.A.; Jockel-Balsarotti, J.; Sullivan, M.; Crisp, M.J.; Kasten, T.; Kirmess, K.M.; et al. Tau Kinetics in Neurons and the Human Central Nervous System. Neuron 2018, 97, 1284–1298. [Google Scholar] [CrossRef] [Green Version]
- Hyman, B.T. Amyloid-Dependent and Amyloid-Independent Stages of Alzheimer Disease. Arch. Neurol. 2011, 68, 1062–1064. [Google Scholar] [CrossRef]
- Stancu, I.-C.; Vasconcelos, B.; Terwel, D.; Dewachter, I. Models of β-amyloid induced Tau-pathology: The long and “folded” road to understand the mechanism. Mol. Neurodegener. 2014, 9, 51–65. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Guo, J.L.; McBride, J.D.; Narasimhan, S.; Kim, H.; Changolkar, L.; Zhang, B.; Gathagan, R.J.; Yue, C.; Dengler, C.; et al. Amyloid-β plaques enhance Alzheimer’s brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat. Med. 2018, 24, 29–38. [Google Scholar] [CrossRef]
- Shin, W.S.; Di, J.; Cao, Q.; Li, B.; Seidler, P.M.; Murray, K.A.; Bitan, G.; Jiang, L. Amyloid β-protein oligomers promote the uptake of tau fibril seeds potentiating intracellular tau aggregation. Alzheimers Res. Ther. 2019, 11, 86–99. [Google Scholar] [CrossRef] [Green Version]
- Shin, W.S.; Di, J.; Murray, K.A.; Sun, C.; Li, B.; Bitan, G.; Jiang, L. Different Amyloid-β Self-Assemblies Have Distinct Effects on Intracellular Tau Aggregation. Front. Mol. Neurosci. 2019, 12, 268–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickett, E.K.; Herrmann, A.G.; McQueen, J.; Abt, K.; Dando, O.; Tulloch, J.; Jain, P.; Dunnett, S.; Sohrabi, S.; Fjeldstad, M.P.; et al. Amyloid Beta and Tau Cooperate to Cause Reversible Behavioral and Transcriptional Deficits in a Model of Alzheimer’s Disease. Cell Rep. 2019, 29, 3592–3604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ising, C.; Venegas, C.; Zhang, S.; Scheiblich, H.; Schmidt, S.V.; Vieira-Saecker, A.; Schwartz, S.; Albasset, S.; McManus, R.M.; Tejera, D.; et al. NLRP3 inflammasome activation drives tau pathology. Nature 2019, 575, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Avila, J.; Schöll, M.; Kovacs, G.G.; Kövari, E.; Skrabana, R.; Evans, L.D.; Kontsekova, E.; Malawska, B.; de Silva, R.; et al. A walk through tau therapeutic strategies. Acta Neuropathol. Commun. 2019, 7, 22–53. [Google Scholar] [CrossRef]
- Morsy, A.; Trippier, P.C. Current and Emerging Pharmacological Targets for the Treatment of Alzheimer’s Disease. J. Alzheimers Dis. 2019, 72, S145–S176. [Google Scholar] [CrossRef]
- Huang, L.-K.; Chao, S.-P.; Hu, C.-J. Clinical trials of new drugs for Alzheimer disease. J. Biomed. Sci. 2020, 27, 18–31. [Google Scholar] [CrossRef]
- Cummings, J.; Lee, G.; Ritter, A.; Sabbagh, M.; Zhong, K. Alzheimer’s disease drug development pipeline: 2019. Alzheimers Dement. Transl. Res. Clin. Interv. 2019, 5, 272–293. [Google Scholar] [CrossRef]
- Hara, Y.; McKeehan, N.; Fillit, H.M. Translating the biology of aging into novel therapeutics for Alzheimer disease. Neurology 2019, 92, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Savinov, S.N.; Fejzo, J.; Eyles, S.J.; Chen, J. Modulation of Amyloid-β42 Conformation by Small Molecules Through Nonspecific Binding. J. Chem. Theory Comput. 2019, 15, 5169–5174. [Google Scholar] [CrossRef]
- Hamlett, E.D.; Hjorth, E.; Ledreux, A.; Gilmore, A.; Schultzberg, M.; Granholm, A.C. RvE1 treatment prevents memory loss and neuroinflammation in the Ts65Dn mouse model of Down syndrome. Glia 2020, 1–14. [Google Scholar] [CrossRef]
- Derakhshankhah, H.; Sajadimajd, S.; Jafari, S.; Izadi, Z.; Sarvari, S.; Sharifi, M.; Falahati, M.; Moakedi, F.; Muganda, W.C.A.; Müller, M.; et al. Novel therapeutic strategies for Alzheimer’s disease: Implications from cell-based therapy and nanotherapy. Nanomed. Nanotechnol. Biol. Med. 2020, 24, 102149–102220. [Google Scholar] [CrossRef] [PubMed]






| Toxic Subpopulation | Non-Toxic Subpopulation |
|---|---|
| Mw > 50 kDa (HMW, high mol. weight) | Mw < 50kDa (LMW, low mol. weight) |
| unrelated to plaques | related to plaques |
| no reaction with anti fAβ antibody | binding anti-fAβ antibody |
| “type 1” AβO: toxic | “type 2” AβO: non-toxic |
| disrupt memory function | no effect on memory function |
| Name | MW (kDa) | Length (AA) | |||
|---|---|---|---|---|---|
| h tau 40 | 45.9 | 441 |  |  | |
| h tau 34 | 43.0 | 412 |  | 4R | |
| h tau 24 | 40.0 | 383 |  | ||
| h tau 39 | 42.6 | 410 |  |  | |
| h tau 37 | 39.7 | 381 |  | 3R | |
| h tau 23 | 36.7 | 352 |  |
| Name | Size | Toxicity |
|---|---|---|
| Tau monomers | 67–70 kDa (352–441 AA) | non-toxic |
| Abnormally phosphorylated monomer | 67–70 kDa | toxic |
| Tau dimer-trimer | 120–180 kDa | toxic |
| Small soluble oligomers (TauOs, 6–8 units) | 300–500 kDa | toxic |
| Granular tau oligomers (gTauOs, 36 units) | 1800 kDa | toxic |
| Straight filaments (SFs) | 50 nm x 10 nm | not always toxic |
| Paired helical filaments (PHFs) | 80 nm x 10–20nm | probably non-toxic |
| Neurofibrillary tangles (NFTs) | “ | probably non-toxic |
| Ghost tangle (originated from degenerated neurons) | probably non-toxic |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penke, B.; Szűcs, M.; Bogár, F. Oligomerization and Conformational Change Turn Monomeric β-Amyloid and Tau Proteins Toxic: Their Role in Alzheimer’s Pathogenesis. Molecules 2020, 25, 1659. https://doi.org/10.3390/molecules25071659
Penke B, Szűcs M, Bogár F. Oligomerization and Conformational Change Turn Monomeric β-Amyloid and Tau Proteins Toxic: Their Role in Alzheimer’s Pathogenesis. Molecules. 2020; 25(7):1659. https://doi.org/10.3390/molecules25071659
Chicago/Turabian StylePenke, Botond, Mária Szűcs, and Ferenc Bogár. 2020. "Oligomerization and Conformational Change Turn Monomeric β-Amyloid and Tau Proteins Toxic: Their Role in Alzheimer’s Pathogenesis" Molecules 25, no. 7: 1659. https://doi.org/10.3390/molecules25071659