Abstract
Histological and molecular analyses of urothelial carcinoma often reveal intratumoural and intertumoural heterogeneity at the genomic, transcriptional and cellular levels. Despite the clonal initiation of the tumour, progression and metastasis often arise from subclones that can develop naturally or during therapy, resulting in molecular alterations with a heterogeneous distribution. Variant histologies in tumour tissues that have developed distinct morphological characteristics divergent from urothelial carcinoma are extreme examples of tumour heterogeneity. Ultimately, heterogeneity contributes to drug resistance and relapse after therapy, resulting in poor survival outcomes. Mutation profile differences between patients with muscle-invasive and metastatic urothelial cancer (interpatient heterogeneity) probably contribute to variability in response to chemotherapy and immunotherapy as first-line treatments. Heterogeneity can occur on multiple levels and averaging or normalizing these alterations is crucial for clinical trial and drug design to enable appropriate therapeutic targeting. Identification of the extent of heterogeneity might shape the choice of monotherapy or additional combination treatments to target different drivers and genetic events. Identification of the lethal tumour cell clones is required to improve survival of patients with urothelial carcinoma.
Key points
-
Bladder cancers have a high total mutational burden and considerable intratumoural and intertumoural heterogeneity at the genomic, transcriptional and cellular levels that remain difficult to quantify.
-
Heterogeneity might be driven by genomic events initiated by APOBEC enzymes and selection pressure from therapeutic interventions, which both drive tumour evolution.
-
Bladder tumours can be categorized into different subtypes on the basis of gene expression signatures, but these molecular subtypes might be unstable and different subtypes can occur within the same tumour causing intratumoural heterogeneity.
-
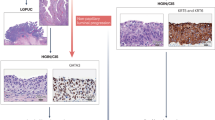
Variant tumour histologies are the morphological extreme of tumour heterogeneity and include glandular, squamous, nested, plasmacytoid, micropapillary, sarcomatoid and small-cell carcinoma.
-
Tumour heterogeneity might affect treatment efficacy, for example, of neoadjuvant chemotherapy and immune checkpoint inhibitors, as well as targeted therapy, for example, when individual actionable mutations only occur in a fraction of the tumour.
-
Biomarkers to select personalized treatments in precision medicine approaches will likely shape future clinical trial design, but their validity might be affected by heterogeneity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Lamy, P. et al. Paired exome analysis reveals clonal evolution and potential therapeutic targets in urothelial carcinoma. Cancer Res. 76, 5894–5906 (2016).
da Costa, J. B. et al. Molecular tumor heterogeneity in muscle invasive bladder cancer: biomarkers, subtypes, and implications for therapy. Urol. Oncol. https://doi.org/10.1016/j.urolonc.2018.11.015 (2018).
Warrick, J. I. et al. Intratumoral heterogeneity of bladder cancer by molecular subtypes and histologic variants. Eur. Urol. 75, 18–22 (2019).
Ma, G. et al. Precision medicine and bladder cancer heterogeneity. Bull. Cancer 105, 925–931 (2018).
Liu, D. et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat. Commun. 8, 2193 (2017).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Russo, M. et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 6, 147–153 (2016).
Marks, E., Rizvi, S. M., Sarwani, N., Yang, Z. & El-Deiry, W. S. A case of heterogeneous sensitivity to panitumumab in cetuximab-refractory colorectal adenocarcinoma metastases. Cancer Biol. Ther. 16, 377–382 (2015).
Pauli, C. et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 7, 462–477 (2017).
Babjuk, M. et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur. Urol. 71, 447–461 (2017).
Dyrskjøt, L. & Ingersoll, M. A. Biology of nonmuscle-invasive bladder cancer: pathology, genomic implications, and immunology. Curr. Opin. Urol. 28, 598–603 (2018).
Hurst, C. D. & Knowles, M. A. Mutational landscape of non-muscle-invasive bladder cancer. Urol. Oncol. https://doi.org/10.1016/j.urolonc.2018.10.015 (2018).
Gerlinger, M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014).
Epstein, J. I. et al. The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am. J. Surg. Pathol. 40, 244–252 (2016).
Faltas, B. M. et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat. Genet. 48, 1490–1499 (2016).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
McPherson, A. et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet. 48, 758–767 (2016).
The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014).
Findlay, J. M. et al. Differential clonal evolution in oesophageal cancers in response to neo-adjuvant chemotherapy. Nat. Commun. 7, 11111 (2016).
McQuerry, J. A., Chang, J. T., Bowtell, D. D. L., Cohen, A. & Bild, A. H. Mechanisms and clinical implications of tumor heterogeneity and convergence on recurrent phenotypes. J. Mol. Med. 95, 1167–1178 (2017).
Meeks, J. J., Goldkorn, A., Aparicio, A. M. & McConkey, D. J. Development of a translational medicine protocol for an NCTN genitourinary clinical trial: critical steps, common pitfalls and a basic guide to translational clinical research. Urol. Oncol. 37, 313–317 (2019).
Sharma, M., Gogia, A., Deo, S. S. V. & Mathur, S. Role of rebiopsy in metastatic breast cancer at progression. Curr. Probl. Cancer 43, 438–442 (2019).
Hench, I. B., Hench, J. & Tolnay, M. Liquid biopsy in clinical management of breast, lung, and colorectal cancer. Front. Med. 5, 9 (2018).
Christensen, E. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
Thierry, A. R. et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann. Oncol. 28, 2149–2159 (2017).
Agarwal, N. et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer 124, 2115–2124 (2018).
Paweletz, C. P. et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin. Cancer Res. 22, 915–922 (2016).
Barata, P. C. et al. Next-generation sequencing (NGS) of cell-free circulating tumor DNA and tumor tissue in patients with advanced urothelial cancer: a pilot assessment of concordance. Ann. Oncol. 28, 2458–2463 (2017).
Pal, S. K. et al. Efficacy of BGJ398, a fibroblast growth factor receptor 1-3 Inhibitor, in patients with previously treated advanced urothelial carcinoma with FGFR3 alterations. Cancer Discov. 8, 812–821 (2018).
Oellerich, M. et al. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 54, 205–218 (2017).
Chalmers, Z. R. et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9, 34 (2017).
Nordentoft, I. et al. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 7, 1649–1663 (2014).
Cha, E. K. et al. Branched evolution and intratumor heterogeneity of urothelial carcinoma of the bladder. J. Clin. Oncol. 32, 293–293 (2014).
Prandi, D. et al. Unraveling the clonal hierarchy of somatic genomic aberrations. Genome Biol. 15, 439 (2014).
Thomsen, M. B. H. et al. Spatial and temporal clonal evolution during development of metastatic urothelial carcinoma. Mol. Oncol. 10, 1450–1460 (2016).
Dentro, S. C., Wedge, D. C. & Van Loo, P. Principles of reconstructing the subclonal architecture of cancers. Cold Spring Harb. Perspect. Med. https://doi.org/10.1101/cshperspect.a026625 (2017).
Waclaw, B. et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 525, 261–264 (2015).
Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540–556 (2018).
Moris, A., Murray, S. & Cardinaud, S. AID and APOBECs span the gap between innate and adaptive immunity. Front. Microbiol. 5, 534 (2014).
Swanton, C., McGranahan, N., Starrett, G. J. & Harris, R. S. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5, 704–712 (2015).
Salter, J. D., Bennett, R. P. & Smith, H. C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 41, 578–594 (2016).
Roberts, S. A. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013).
Burns, M. B., Temiz, N. A. & Harris, R. S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45, 977–983 (2013).
Hedegaard, J. et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 30, 27–42 (2016).
Blanes, A., Rubio, J., Sanchez-Carrillo, J. J. & Diaz-Cano, S. J. Coexistent intraurothelial carcinoma and muscle-invasive urothelial carcinoma of the bladder: clonality and somatic down-regulation of DNA mismatch repair. Hum. Pathol. 40, 988–997 (2009).
Kazanov, M. D. et al. APOBEC-induced cancer mutations are uniquely enriched in early-replicating, gene-dense, and active chromatin regions. Cell Rep. 13, 1103–1109 (2015).
Hoopes, J. I. et al. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 14, 1273–1282 (2016).
Seplyarskiy, V. B. et al. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res. 26, 174–182 (2016).
Nowarski, R. & Kotler, M. APOBEC3 cytidine deaminases in double-strand DNA break repair and cancer promotion. Cancer Res. 73, 3494–3498 (2013).
Okeoma, C. M., Lovsin, N., Peterlin, B. M. & Ross, S. R. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445, 927–930 (2007).
Smith, H. C., Bennett, R. P., Kizilyer, A., McDougall, W. M. & Prohaska, K. M. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 23, 258–268 (2012).
Law, E. K. et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2, e1601737 (2016).
Sjödahl, G. et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 18, 3377–3386 (2012).
Choi, W. et al. Genetic alterations in the molecular subtypes of bladder cancer: illustration in the cancer genome atlas dataset. Eur. Urol. 72, 354–365 (2017).
Kamoun, A. et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur. Urol. 77, 420–433 (2020).
Choi, W. et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 25, 152–165 (2014).
Seiler, R. et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur. Urol. 72, 544–554 (2017).
Mariathasan, S. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018).
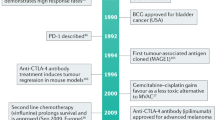
Rosenberg, J. E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016).
Sharma, P. et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 18, 312–322 (2017).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Rousseeuw, P. J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987).
Dyrskjøt, L. et al. A molecular signature in superficial bladder carcinoma predicts clinical outcome. Clin. Cancer Res. 11, 4029–4036 (2005).
Dyrskjøt, L. et al. Prognostic impact of a 12-gene progression score in non-muscle-invasive bladder cancer: a prospective multicentre validation study. Eur. Urol. 72, 461–469 (2017).
Sjödahl, G. et al. Discordant molecular subtype classification in the basal-squamous subtype of bladder tumors and matched lymph-node metastases. Mod. Pathol. 31, 1869–1881 (2018).
Thomsen, M. B. H. et al. Comprehensive multiregional analysis of molecular heterogeneity in bladder cancer. Sci. Rep. 7, 11702 (2017).
Ma, S. et al. Continuity of transcriptomes among colorectal cancer subtypes based on meta-analysis. Genome Biol. 19, 142 (2018).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018).
Jerby-Arnon, L. et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997.e24 (2018).
Amin, M. B. Histological variants of urothelial carcinoma: diagnostic, therapeutic and prognostic implications. Mod. Pathol. 22, S96–S118 (2009).
Linder, B. J. et al. The impact of histological reclassification during pathology re-review — evidence of a Will Rogers effect in bladder cancer? J. Urol. 190, 1692–1696 (2013).
Shah, R. B., Montgomery, J. S., Montie, J. E. & Kunju, L. P. Variant (divergent) histologic differentiation in urothelial carcinoma is under-recognized in community practice: impact of mandatory central pathology review at a large referral hospital. Urol. Oncol. 31, 1650–1655 (2013).
Moch, H., Humphrey, P. A., Ulbright, T. M. & Reuter, V. E. WHO Classification of Tumours of the Urinary System and Male Genital Organs Vol. 4, 77–133 (International Agency for Research on Cancer, 2016).
Humphrey, P. A., Moch, H., Cubilla, A. L., Ulbright, T. M. & Reuter, V. E. The 2016 WHO classification of tumours of the urinary system and male genital organs — part B: prostate and bladder tumours. Eur. Urol. 70, 106–119 (2016).
Hovelson, D. H. et al. Targeted DNA and RNA sequencing of paired urothelial and squamous bladder cancers reveals discordant genomic and transcriptomic events and unique therapeutic implications. Eur. Urol. 74, 741–753 (2018).
Blochin, E. B., Park, K. J., Tickoo, S. K., Reuter, V. E. & Al-Ahmadie, H. Urothelial carcinoma with prominent squamous differentiation in the setting of neurogenic bladder: role of human papillomavirus infection. Mod. Pathol. 25, 1534–1542 (2012).
Chapman-Fredricks, J. R. et al. High-risk human papillomavirus DNA detected in primary squamous cell carcinoma of urinary bladder. Arch. Pathol. Lab. Med. 137, 1088–1093 (2013).
Solomon, J. P. et al. Challenges in the diagnosis of urothelial carcinoma variants: can emerging molecular data complement pathology review? Urology 102, 7–16 (2016).
Vail, E. et al. Telomerase reverse transcriptase promoter mutations in glandular lesions of the urinary bladder. Ann. Diagn. Pathol. 19, 301–305 (2015).
Lopez-Beltran, A. et al. Variants and new entities of bladder cancer. Histopathology 74, 77–96 (2019).
Dhall, D., Al-Ahmadie, H. & Olgac, S. Nested variant of urothelial carcinoma. Arch. Pathol. Lab. Med. 131, 1725–1727 (2007).
Volmar, K. E., Chan, T. Y., De Marzo, A. M. & Epstein, J. I. Florid von Brunn nests mimicking urothelial carcinoma: a morphologic and immunohistochemical comparison to the nested variant of urothelial carcinoma. Am. J. Surg. Pathol. 27, 1243–1252 (2003).
Zhong, M. et al. Distinguishing nested variants of urothelial carcinoma from benign mimickers by TERT promoter mutation. Am. J. Surg. Pathol. 39, 127–131 (2015).
Al-Ahmadie, H. A. et al. Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat. Genet. 48, 356–358 (2016).
Keck, B. et al. The plasmacytoid carcinoma of the bladder–rare variant of aggressive urothelial carcinoma. Int. J. Cancer 129, 346–354 (2011).
Nigwekar, P. et al. Plasmacytoid urothelial carcinoma: detailed analysis of morphology with clinicopathologic correlation in 17 cases. Am. J. Surg. Pathol. 33, 417–424 (2009).
Dayyani, F. et al. Plasmacytoid urothelial carcinoma, a chemosensitive cancer with poor prognosis, and peritoneal carcinomatosis. J. Urol. 189, 1656–1661 (2013).
Kaimakliotis, H. Z. et al. Plasmacytoid bladder cancer: variant histology with aggressive behavior and a new mode of invasion along fascial planes. Urology 83, 1112–1116 (2014).
Amin, M. B. et al. Micropapillary variant of transitional cell carcinoma of the urinary bladder. Histologic pattern resembling ovarian papillary serous carcinoma. Am. J. Surg. Pathol. 18, 1224–1232 (1994).
Nassar, H. et al. Pathogenesis of invasive micropapillary carcinoma: role of MUC1 glycoprotein. Mod. Pathol. 17, 1045–1050 (2004).
Luna-More, S., Gonzalez, B., Acedo, C., Rodrigo, I. & Luna, C. Invasive micropapillary carcinoma of the breast. A new special type of invasive mammary carcinoma. Pathol. Res. Pract. 190, 668–674 (1994).
Ching, C. B. et al. HER2 gene amplification occurs frequently in the micropapillary variant of urothelial carcinoma: analysis by dual-color in situ hybridization. Mod. Pathol. 24, 1111–1119 (2011).
Ross, J. S. et al. A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clin. Cancer Res. 20, 68–75 (2014).
Isharwal, S. et al. Intratumoral heterogeneity of ERBB2 amplification and HER2 expression in micropapillary urothelial carcinoma. Hum. Pathol. 77, 63–69 (2018).
Iyer, G. et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J. Clin. Oncol. 31, 3133–3140 (2013).
Fleischmann, A., Rotzer, D., Seiler, R., Studer, U. E. & Thalmann, G. N. Her2 amplification is significantly more frequent in lymph node metastases from urothelial bladder cancer than in the primary tumours. Eur. Urol. 60, 350–357 (2011).
Lopez-Beltran, A. et al. Carcinosarcoma and sarcomatoid carcinoma of the bladder: clinicopathological study of 41 cases. J. Urol. 159, 1497–1503 (1998).
Sanfrancesco, J. et al. Sarcomatoid urothelial carcinoma of the bladder: analysis of 28 cases with emphasis on clinicopathologic features and markers of epithelial-to-mesenchymal transition. Arch. Pathol. Lab. Med. 140, 543–551 (2016).
Sung, M. T. et al. Histogenesis of sarcomatoid urothelial carcinoma of the urinary bladder: evidence for a common clonal origin with divergent differentiation. J. Pathol. 211, 420–430 (2007).
Guo, C. C. et al. Dysregulation of EMT drives the progression to clinically aggressive sarcomatoid bladder cancer. Cell Rep. 27, 1781–1793.e4 (2019).
George, J. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015).
Chang, M. T. et al. Small cell carcinomas of the bladder and lung are characterized by a convergent but distinct pathogenesis. Clin. Cancer Res. 24, 1965–1973 (2018).
Kim, P. H. et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur. Urol. 67, 198–201 (2015).
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016).
Sjödahl, G., Eriksson, P., Liedberg, F. & Höglund, M. Molecular classification of urothelial carcinoma: global mRNA classification versus tumour-cell phenotype classification. J. Pathol. 242, 113–125 (2017).
Batista da Costa, J. et al. Molecular characterization of neuroendocrine-like bladder cancer. Clin. Cancer Res. 25, 3908–3920 (2019).
Gopalan, A. et al. Urachal carcinoma: a clinicopathologic analysis of 24 cases with outcome correlation. Am. J. Surg. Pathol. 33, 659–668 (2009).
Jordan, E. et al. Assessment of genomic alterations in bladder adenocarcinoma and urachal adenocarcinoma. Eur. J. Cancer 51, S530–S530 (2015).
Collazo-Lorduy, A. et al. Urachal carcinoma shares genomic alterations with colorectal carcinoma and may respond to epidermal growth factor inhibition. Eur. Urol. 70, 771–775 (2016).
Reis, H. et al. Pathogenic and targetable genetic alterations in 70 urachal adenocarcinomas. Int. J. Cancer 143, 1764–1773 (2018).
Saunders, N. A. et al. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol. Med. 4, 675–684 (2012).
Garraway, L. A. & Janne, P. A. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2, 214–226 (2012).
Steinbichler, T. B. et al. Therapy resistance mediated by cancer stem cells. Semin. Cancer Biol. 53, 156–167 (2018).
Guo, C. C. et al. Gene expression profile of the clinically aggressive micropapillary variant of bladder cancer. Eur. Urol. 70, 611–620 (2016).
Meeks, J. J. et al. A systematic review of neoadjuvant and adjuvant chemotherapy for muscle-invasive bladder cancer. Eur. Urol. 62, 523–533 (2012).
Grossman, H. B. et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N. Engl. J. Med. 349, 859–866 (2003).
Plimack, E. R. et al. Accelerated methotrexate, vinblastine, doxorubicin, and cisplatin is safe, effective, and efficient neoadjuvant treatment for muscle-invasive bladder cancer: results of a multicenter phase II study with molecular correlates of response and toxicity. J. Clin. Oncol. 32, 1895–1901 (2014).
Kamat, A. M. et al. Bladder cancer. Lancet 388, 2796–2810 (2016).
Lotan, Y. et al. Molecular subtyping of clinically localized urothelial carcinoma reveals lower rates of pathological upstaging at radical cystectomy among luminal tumors. Eur. Urol. 76, 200–206 (2019).
The ASCO Post. FDA grants accelerated approval to erdafitinib for metastatic urothelial carcinoma. The ASCO Post https://www.ascopost.com/News/59934 (2019).
Loriot, Y. et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 381, 338–348 (2019).
Pouessel, D. et al. Tumor heterogeneity of fibroblast growth factor receptor 3 (FGFR3) mutations in invasive bladder cancer: implications for perioperative anti-FGFR3 treatment. Ann. Oncol. 27, 1311–1316 (2016).
Goldstein, J. T. et al. Genomic activation of PPARG reveals a candidate therapeutic axis in bladder cancer. Cancer Res. 77, 6987–6998 (2017).
Siefker-Radtke, A. & Curti, B. Immunotherapy in metastatic urothelial carcinoma: focus on immune checkpoint inhibition. Nat. Rev. Urol. 15, 112–124 (2018).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949.e16 (2017).
Necchi, A. et al. Pembrolizumab as neoadjuvant therapy before radical cystectomy in patients with muscle-invasive urothelial bladder carcinoma (PURE-01): an open-label, single-arm, phase II study. J. Clin. Oncol. 6, 3353–3360 (2018).
US National Library of Medicine. ClinicalTrials.gov http://www.clinicaltrials.gov/ct2/show/NCT02177695 (2019).
Flaig, T. W. et al. SWOG S1314: a randomized phase II study of co-expression extrapolation (COXEN) with neoadjuvant chemotherapy for localized, muscle-invasive bladder cancer. J. Clin. Oncol. 37, 4506–4506 (2019).
Christensen, E. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
Hurst, C. D. et al. Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer Cell 32, 701–715.e7 (2017).
Vandekerkhove, G. et al. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin. Cancer Res. 23, 6487–6497 (2017).
Acknowledgements
The authors acknowledge the Leo and Anne Albert Institute of Bladder Cancer Research. J.J.M. is supported by a VA Merit Award (BX003692-01) and a Department of Defense Grant (W81XWH-18-0257). J.J.M and L.D. are supported by an award from the Leo and Anne Albert Institute of Bladder Cancer Research. H.A. is supported by a Sloan Kettering Institute for Cancer Research Cancer Center Support Grant P30CA008748 and SPORE in Bladder Cancer P50CA221745. B.M.F. is supported by the Department of Defense CDMRP Career Development Award (grant CA160212). D.J.D. is supported in part by RSG 17-233-01-TBE from the American Cancer Society. B.L.W. is supported by an American Urological Association Research Scholar Award.
Author information
Authors and Affiliations
Contributions
J.J.M., H.A., B.M.F., J.A.T.III, T.W.F., E.C., B.L.W., D.J.M., L.D. researched data for the article and wrote the manuscript. All authors made substantial contributions to discussion of the article content and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
B.M.F. received grant/research support from Eli Lilly and Company and participated in an Advisory Board for Immunomedics. J.J.M. received research support from Epizyme, Tesaro and Abbvie, is a consultant for Ferring and AstraZeneca, and participated in advisory boards for Cold Genesys and Janssen. H.A. is a consultant for AstraZeneca/MedImmune, Bristol-Myers Squibb and EMD Serono. L.D. received research support from Ferring and Natera, and is a consultant for Ferring.
Additional information
Peer review information
Nature Reviews Urology thanks P. Black, R. Bryan and A. Hartmann for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Meeks, J.J., Al-Ahmadie, H., Faltas, B.M. et al. Genomic heterogeneity in bladder cancer: challenges and possible solutions to improve outcomes. Nat Rev Urol 17, 259–270 (2020). https://doi.org/10.1038/s41585-020-0304-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41585-020-0304-1
This article is cited by
-
The sex gap in bladder cancer survival — a missing link in bladder cancer care?
Nature Reviews Urology (2024)
-
Sakuranin represses the malignant biological behaviors of human bladder cancer cells by triggering autophagy via activating the p53/mTOR pathway
BMC Urology (2023)
-
Comparative proteomics analysis in different stages of urothelial bladder cancer for identification of potential biomarkers: highlighted role for antioxidant activity
Clinical Proteomics (2023)
-
Optimizing identification of consensus molecular subtypes in muscle-invasive bladder cancer: a comparison of two sequencing methods and gene sets using FFPE specimens
BMC Cancer (2023)
-
Transcriptional state dynamics lead to heterogeneity and adaptive tumor evolution in urothelial bladder carcinoma
Communications Biology (2023)