Abstract
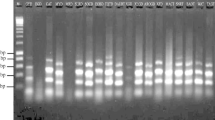
Drumstick (Moringa oleifera Lam.) is an important vegetable as well as forage crop of arid and semi-arid zones of the tropics. The leaves and pods of the plant are rich sources of minerals and vitamins. In the present work, genetic diversity study of 23 genotypes of M. oleifera collected from Kerala, Tamil Nadu and Karnataka states of India was carried out using seven cytochrome P450 (CytP450) markers. By using seven pairs of CytP450 gene-based markers, 88.25% of polymorphism was recorded among the 23 sampled genotypes. The Polymorphic Information Content (PI), Marker Index (MI) and Resolving Power obtained for seven primers were estimated 0.23, 2.96 and 9.83, respectively. The Unweighted Pair Group Method with Arithmetic mean (UPGMA) dendrogram based on this marker data indicate that genotypes from different geographical regions are placed in the same clusters. The dendrogram and Principal Coordinates Analysis (PCoA) plots derived from the binary data matrices were highly concordant. The investigation, in brief, proved that CytP450 based marker system is efficient in the elucidation of genetic diversity in M. oleifera accessions.




Similar content being viewed by others
References
Olson ME (2002) Combining data from DNA sequences and morphology for a phylogeny of moringaceae (Brassicales). Syst Bot 27:55–73. https://doi.org/10.1043/0363-6445-27.1.55
Parrotta JA (2009) Moringa oleifera LAM, 1785. Enzyklopädie der Holzgewachse, Handb und Atlas der Dendrol. https://doi.org/10.1089/jop.2010.0049
Saini RK, Saad KR, Ravishankar GA, Giridhar P, Shetty NP (2013) Genetic diversity of commercially grown Moringa oleifera Lam. cultivars from India by RAPD, ISSR and cytochrome P450-based markers. Plant Syst Evol 299:1205–1213. https://doi.org/10.1007/s00606-013-0789-7
Muluvi GM, Sprent JI, Soranzo N, Provan J, Odee D, Folkard G, McNicol JW, Powell W (1999) Amplified fragment length polymorphism (AFLP) analysis of genetic variation in Moringa oleifera Lam. Mol Ecol 8:463–470. https://doi.org/10.1046/j.1365-294X.1999.00589.x
Nambiar VS, Bhadalkar K, Daxini M (2003) Drumstick leaves as source of vitamin A in ICDS-SFP. Indian J Pediatr 70:383–387. https://doi.org/10.1007/BF02723611
Oduro I, Ellis WO, Owusu D (2008) Nutritional potential of two leafy vegetables : Moringa oleifera and Ipomoea batatas leaves. Sci Res Essay 3:57–60. https://doi.org/10.1016/S0167-8809(00)00138-9
Dalia I, Machado S, José A, Gastélum N, Moreno CR, Wong BR, Cervantes JL (2010) Nutritional quality of edible parts of Moringa oleifera. Food Anal Methods 3:175–180. https://doi.org/10.1007/s12161-009-9106-z
Witt K (2014) The nutrient content of Moringa oleifera Leaves. Echo Research Note No. 1. https://doi.org/10.4135/9781849209984.n1
Chen G, Yang M, Kuo P, Lin M (2014) Chemical constituents of Moringa oleifera and their cytotoxicity aganist doxorubicin-resistant human. Chem Nat Compd 50:154–156
Abalaka EM, Daniyan YS, Oyeleke BS, Adeyemo OS (2012) The Antibacterial evaluation of Moringa Oleifera leaf extracts on selected bacterial pathogens. J Microbiol Res 2:1–4. https://doi.org/10.5923/j.microbiology.20120202.01
Chuang HP, Lee CW, Chou JY, Murugan M, Shieh BJ, Chen HM (2007) Anti-fungal activity of crude extracts and essential oil of Moringa oleifera Lam. Bioresour Technol 98:232–236. https://doi.org/10.1016/j.biortech.2005.11.003
Sreelatha S, Padma PR (2009) Antioxidant activity and total phenolic content of Moringa oleifera leaves in two stages of maturity. Plant Foods Hum Nutr 64:303–311. https://doi.org/10.1007/s11130-009-0141-0
Bekele B, Adane L, Tariku Y, Hailu A (2013) Evaluation of antileishmanial activities of triglycerides isolated from roots of Moringa stenopetala. Med Chem Res 22:4592–4599. https://doi.org/10.1007/s00044-013-0467-x
Ghasi S, Nwobodo E, Ofili JO (2000) Hypocholesterolemic effects of crude extract of leaf of Moringa oleifera Lam. in high-fat diet fed wistar rats. J Ethnopharmacol 69:21–25
Poczai P, Varga I, Bell NE, Hyvönen J (2012) Genomics Meets Biodiversity: Advances in Molecular Marker Development and Their Applications in Plant Genetic Diversity Assessment. J Genomics Meets Biodivers. https://doi.org/10.5772/33614
Yadav S, Srivastava J (2014) of Recent scientific research article gentic diversity analysis on Moringa oliefera by using different molecular markers: a review. Int J Recent Sci Res 5:2277–2282
Zargar SM, Farhat S, Mahajan R, Bhakri A, Sharma A (2016) Unraveling the efficiency of RAPD and SSR markers in diversity analysis and population structure estimation in common bean. Saudi J Biol Sci 23:139–149. https://doi.org/10.1016/j.sjbs.2014.11.011
Fuentes JL, Escobar F, Alvarez A, Gallego G, Duque MC, Ferrer M, Deus JE, Tohme JM (1999) Analysis of genetic diversity in cuban rice varieties using isoenzyme, RAPD and AFLP markers. Euphytica 109:107–115
Sunar S, Yildirim N, Sengul M, Agar G (2016) Genetic diversity and relationships detected by ISSR and RAPD analysis among Aethionema species growing in Eastern Anatolia (Turkey). Comptes Rendus Biol 339:147–151. https://doi.org/10.1016/j.crvi.2016.02.006
Panwar P, Nath M, Yadav VK, Kumar A (2010) Comparative evaluation of genetic diversity using RAPD, SSR and cytochrome P450 gene based markers with respect to calcium content in finger millet (Eleusine coracana L. Gaertn.). J Genet 89:121–133. https://doi.org/10.1007/s12041-010-0052-8
Godwin MM, Katambo MM, Morris NA (2010) Genetic diversity between cultivated and non-cultivated Moringa oleifera Lam. provenances assessed by RAPD markers. J Cell Mol Biol 8:95–102
Ojuederie OB, Igwe DO, Okuofu SI, Faloye B (2013) Assessment of genetic diversity in some Moringa oleifera Lam. landraces from Western Nigeria using RAPD Markers. Afr J Plant Sci Biotechnol 7(1):15–20
Singh S, Kartikeyan K, Singh DR, Sihmachalam P, Biansla NK, Jaisankar I (2017) Genetic diversity in drumstick of Andaman Islands and their relatedness with probable introduction sites from mainland India. Proc Natl Acad Sci India Sect B Biol Sci. https://doi.org/10.1007/s40011-017-0950-0
Kumar GS, Singh R, Choudhury RD, Bharadwaj J, Gupta V, Singode A (2014) Genetic diversity and population structure study of drumstick (Moringa oleifera Lam.) using morphological and SSR markers. Ind Crops Prod 60:316–325. https://doi.org/10.1016/j.indcrop.2014.06.033
Schalk M, Nedelkina S, Schoch G, Batard Y, Reichhart DW (1999) Role of unusual amino acid residues in the proximal and distal heme regions of a plant P450, CYP73A1. Biochem 38:6093–6103. https://doi.org/10.1021/bi982989w
Ohkawa H, Imaishi H, Shiota N, Yamada T, Inui H, Ohkawa Y (1998) Molecular mechanisms of herbicide resistance with special emphasis on cytochrome P450 monooxygenases. Plant Biotechnol 15:173–176
Hallahan DL, West JM (1995) Cytochrome P-450 in plant/insect interactions: geraniol 10-hydroxylase and the biosynthesis of iridoid monoterpenoids. Drug Metabol Drug Interact 12:369–382. https://doi.org/10.1515/DMDI.1995.12.3-4.369
Katoch M, Hussain MA, Ahuja A (2013) Comparison of SSR and cytochrome P-450 markers for estimating genetic diversity in Picrorhiza kurrooa L. Plant Syst Evol 299:1637–1643. https://doi.org/10.1007/s00606-013-0820-z
Tanksley SD, Mccouch SR (1997) Seed unlocking banks and molecular maps: the from potential Wil. Science 277:1063–1066
Somerville C, Somerville S (1999) Plant functional genomics. Science 285:380–383. https://doi.org/10.1126/SCIENCE.285.5426.380
Kumar P, Dolkar R, Manjunatha G, Pallavi HM (2017) Molecular fingerprinting and assessment of genetic variations among advanced breeding lines of Moringa oleifera L. by using seed protein, RAPD and cytochrome P450 based markers. S Afr J Bot 111:60–67. https://doi.org/10.1016/j.sajb.2017.03.024
Saini RK, Shetty NP, Giridhar P, Ravishankar GA (2012) Rapid in vitro regeneration method for Moringa oleifera and performance evaluation of field grown nutritionally enriched tissue cultured plants. 3 Biotech 2:187–192. https://doi.org/10.1007/s13205-012-0045-9
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: Version II. Plant Mol Biol Rep 1:19–22
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Raalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238. https://doi.org/10.1007/BF00564200
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112. https://doi.org/10.1007/s001220051046
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Jaccard P (1908) Nouvelles recherches sur la distribuition florale. Bull Soc Vaud Sci Nat 44:223–270
Rohlf JF (2018) NTSYS-pc numerical taxonomy and multivariate analysis system, Version 2.0, userguide. https://wenku.baidu.com/view/fa1cd829ed630b1c59eeb5f9. Accessed 28 July 2019
Manoko MLK, van den Berg RG, Feron RMC, Van der Weerden GM, Mariani C (2008) Genetic diversity of the African hexaploid species Solanum scrabrum Mill. and Solanum nigrum L. (Solanaceae). Genet Resour Crop Evol 55(3):409–418. https://doi.org/10.1007/s10722-007-9248-z
Panwar P, Saini RK, Sharma N, Yadav D, Kumar A (2010) Efficiency of RAPD, SSR and Cytochrome P450 gene based markers in accessing genetic variability amongst finger millet (Eleusine coracana) accessions. Mol Biol Rep 37:4075–4082. https://doi.org/10.1007/s11033-010-0067-5
Ahmad SM, Hoot SB, Qazi PH, Verma V (2009) Phylogenetic patterns and genetic diversity of Indian Tinospora species based on chloroplast sequence data and cytochrome P450 polymorphisms. Plant Syst Evol 281:87–96. https://doi.org/10.1007/s00606-009-0189-1
Yamanaka S, Suzuki E, Tanaka M, Takeda Y, Watanabe JA, Watanabe KN (2003) Assessment of cytochrome P450 sequences offers a useful tool for determining genetic diversity in higher plant species. Theor Appl Genet 108:1–9. https://doi.org/10.1007/s00122-003-1403-0
Kumar J, Verma V, Qazi GN, Gupta PK (2007) Genetic diversity in Cymbopogon species using PCR-based functional markers. J Plant Biochem Biotechnol 16:119–122
Grativol C, Da Fonseca L-M, Hemerly AS, Ferreira PCG (2011) High efficiency and reliability of inter-simple sequence repeats (ISSR) markers for evaluation of genetic diversity in Brazilian cultivated Jatropha curcas L. accessions. Mol Biol Rep 38:4245–4256. https://doi.org/10.1007/s11033-010-0547-7
Lawrence MJ (2000) Population genetics of the homomorphic self-incompatibility polymorphisms in flowering plants. Ann Bot 85:221–226. https://doi.org/10.1006/anbo.1999.1044
Mathur P, Habibi N, Chittora M, Purohit SD (2013) Molecular analysis of variability among genotypes of Abrus precatorius L. with different seed coat colours using RAPD and ISSR markers. Indian J Biotechnol 12:273–276
Saini M, Singh S, Hussain Z, Yadav A (2010) RAPD analysis in mungbean [Vigna radiata (L.) Wilczek]: I. Assessment of genetic diversity. Indian J Biotechnol 9:137–146
Sindhu A, Tehlan SK, Chaudhury A (2017) Analysis of genetic diversity among medicinal therapist Trigonella foenum-graecum L. genotypes through RAPD and SSR markers. Acta Physiol Plant 39:100. https://doi.org/10.1007/s11738-017-2395-8
Noormohammadi Z, Fasihee A, Rashidpoor SH, Sheidai M, Baraki SG, Mazooji A, Tabatabaee-Ardakani SZ (2012) Genetic variation among Iranian pomegranates (Punica granatum L.) using RAPD, ISSR and SSR markers. Aust J Crop Sci 6:268–275
Rajalakshmi R, Rajalakshmi S, Parida A (2017) Evaluation of the genetic diversity and population structure in drumstick (Moringa oleifera L.) using SSR markers. Curr Sci 112:1–7
Acknowledgements
The authors are grateful to Dr. Suhara Beevy S, Professor and Head, Department of Botany, University of Kerala for the facilities provided. DRRS wish to thank the University of Kerala for granting research fellowship (No.Ac.EI/A2/10625/2016-I) to undertake the present work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ravi, R.S.D., Siril, E.A. & Nair, B.R. The efficiency of Cytochrome P450 gene-based markers in accessing genetic variability of drumstick (Moringa oleifera Lam.) accessions. Mol Biol Rep 47, 2929–2939 (2020). https://doi.org/10.1007/s11033-020-05391-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05391-w