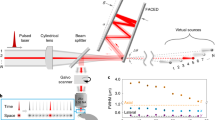
Abstract
Fluorescence is a powerful means to probe information processing in the mammalian brain1. However, neuronal tissues are highly heterogeneous and thus opaque to light. A wide set of non-invasive or invasive techniques for scattered light rejection, optical sectioning or localized excitation have been developed, but non-invasive optical recording of activity through a highly scattering layer beyond the ballistic regime is impossible as yet. Here, we show that functional signals from fluorescent time-varying sources located below a highly scattering bone tissue can be retrieved efficiently by exploiting matrix factorization algorithms to demix this information from temporal sequences of low-contrast fluorescence speckle patterns.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Code availability
Analysis scripts are available at https://github.com/laboGigan/SpeckledNeuronsAnalysis. Hardware control scripts are available at https://github.com/laboGigan/SpeckledNeuronsControl.
References
Helmchen, F. & Konnerth, A. Imaging in Neuroscience: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2011).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Weisenburger, S. & Vaziri, A. A guide to emerging technologies for large-scale and whole-brain optical imaging of neuronal activity. Annu. Rev. Neurosci. 41, 431–452 (2018).
Prevedel, R. et al. Fast volumetric calcium imaging across multiple cortical layers using sculpted light. Nat. Methods 13, 1021–1028 (2016).
Iyer, V., Hoogland, T. M. & Saggau, P. Fast functional imaging of single neurons using random-access multiphoton (RAMP) microscopy. J. Neurophysiol. 95, 535–545 (2006).
Katona, G. et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods 9, 201–208 (2012).
Grewe, B. F., Langer, D., Kasper, H., Kampa, B. M. & Helmchen, F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat. Methods 7, 399–405 (2010).
Bovetti, S. et al. Simultaneous high-speed imaging and optogenetic inhibition in the intact mouse brain. Sci. Rep. 7, 40041 (2017).
Nikolenko, V. SLM microscopy: scanless two-photon imaging and photostimulation using spatial light modulators. Front. Neural Circuits 2, 5 (2008).
Zhou, P. et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728 (2018).
Nöbauer, T. et al. Video rate volumetric Ca2+ imaging across cortex using seeded iterative demixing (SID) microscopy. Nat. Methods 14, 811–818 (2017).
Pégard, N. C. et al. Compressive light-field microscopy for 3D neural activity recording. Optica 3, 517–524 (2016).
Horstmeyer, R., Ruan, H. & Yang, C. Guidestar-assisted wavefront-shaping methods for focusing light into biological tissue. Nat. Photon. 9, 563–571 (2015).
Rotter, S. & Gigan, S. Light fields in complex media: mesoscopic scattering meets wave control. Rev. Mod. Phys. 89, 015005 (2017).
Katz, O., Heidmann, P., Fink, M. & Gigan, S. Non-invasive single-shot imaging through scattering layers and around corners via speckle correlations. Nat. Photon. 8, 784–790 (2014).
Hofer, M., Soeller, C., Brasselet, S. & Bertolotti, J. Wide field fluorescence epi-microscopy behind a scattering medium enabled by speckle correlations. Opt. Express 26, 9866–9881 (2018).
Chang, J. & Wetzstein, G. Single-shot speckle correlation fluorescence microscopy in thick scattering tissue with image reconstruction priors. J. Biophotonics 11, e201700224 (2018).
Stern, G. & Katz, O. Noninvasive focusing through scattering layers using speckle correlations. Opt. Letters 44, 143–146 (2019).
Xu, X. et al. Imaging of objects through a thin scattering layer using a spectrally and spatially separated reference. Opt. Express 26, 15073–15083 (2018).
Judkewitz, B., Horstmeyer, R., Vellekoop, I. M., Papadopoulos, I. N. & Yang, C. Translation correlations in anisotropically scattering media. Nat. Phys. 11, 684–689 (2015).
Hong, G. & Lieber, C. M. Novel electrode technologies for neural recordings. Nat. Rev. Neurosci. 20, 330–345 (2019).
Lin, M. Z. & Schnitzer, M. J. Genetically encoded indicators of neuronal activity. Nat. Neurosci. 19, 1142–1153 (2016).
Deneux, T. et al. Accurate spike estimation from noisy calcium signals for ultrafast three-dimensional imaging of large neuronal populations in vivo. Nat. Commun. 7, 12190 (2016).
Soleimanzad, H., Gurden, H. & Pain, F. Optical properties of mice skull bone in the 455 to 705 nm range. J. Biomed. Opt. 22, 010503 (2017).
Goodman, J. W. Speckle Phenomena in Optics: Theory and Applications (Roberts & Company, 2007).
Comon, P. & Jutten, C. Handbook of Blind Source Separation (Academic Press, 2010).
Mukamel, E. A., Nimmerjahn, A. & Schnitzer, M. J. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760 (2009).
Diego Andilla, F. & Hamprecht, F. A. Sparse space-time deconvolution for calcium image analysis. In Advances in Neural Information Processing Systems 27 (eds Ghahramani, Z. et al.) 64–72 (Curran Associates, 2014).
Maruyama, R. et al. Detecting cells using non-negative matrix factorization on calcium imaging data. Neural Netw. 55, 11–19 (2014).
Pnevmatikakis, E. et al. Simultaneous denoising, deconvolution and demixing of calcium imaging data. Neuron 89, 285–299 (2016).
Mertz, J. & Barankov, R. Imaging luminous objects through a single optical fiber. In Proceedings of Optics in the Life Sciences (2015) BT2A.1 (Optical Society of America, 2015).
Saade, A. et al. Random projections through multiple optical scattering: approximating kernels at the speed of light. In 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 6215–6219 (IEEE, 2016).
Rahimi, A. & Recht, B. Random features for large-scale kernel machines. In Proceeedings of Advances in Neural Information Processing Systems 20 (eds Platt, J. C. et al.) 1177–1184 (Curran Associates, 2008).
Mahoney, M. in Foundations and Trends in Machine Learning Vol. 3, 123–224 (Now Publishers, 2011).
Boniface, A., Blochet, B., Dong, J. & Gigan, S. Noninvasive light focusing in scattering media using speckle variance optimization. Optica 6, 1381–1385 (2019).
Acknowledgements
We thank L. Bourdieu for providing biological samples and for numerous discussions, and F. Niwa for the cultured neurons. We also thank A. Vaziri and T. Nöbauer for useful suggestions, S. Leedumrongwatthanakun, J. Dong and A. Boniface for constructive comments, and B. Rauer and F. Soldevila for comments on the manuscript. This work was funded by the European Research Council (ERC; H2020, SMARTIES-724473). S.G. is a member of the Institut Universitaire de France.
Author information
Authors and Affiliations
Contributions
C.M. performed the experiment and analysed the data. S.G. and C.M. conceived the project and wrote the manuscript. S.G. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods and Figs. 1–13.
Rights and permissions
About this article
Cite this article
Moretti, C., Gigan, S. Readout of fluorescence functional signals through highly scattering tissue. Nat. Photonics 14, 361–364 (2020). https://doi.org/10.1038/s41566-020-0612-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41566-020-0612-2
This article is cited by
-
Overlapping speckle correlation algorithm for high-resolution imaging and tracking of objects in unknown scattering media
Nature Communications (2023)
-
Robust and adjustable dynamic scattering compensation for high-precision deep tissue optogenetics
Communications Biology (2023)
-
Consequences of nano and microplastic exposure in rodent models: the known and unknown
Particle and Fibre Toxicology (2022)
-
Large field-of-view non-invasive imaging through scattering layers using fluctuating random illumination
Nature Communications (2022)
-
Deep tissue localization and sensing using optical microcavity probes
Nature Communications (2022)