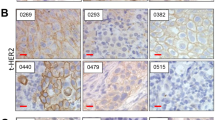
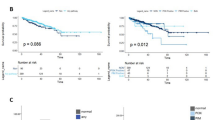
Abstract
Muscle-invasive bladder carcinomas (MIBCs) are aggressive genitourinary malignancies. Metastatic urothelial carcinoma of the bladder is generally incurable by current chemotherapy and leads to early mortality. Recent studies have identified molecular subtypes of MIBCs with different sensitivities to frontline therapy, suggesting tumor heterogeneity. We have performed multi-omic profiling of the kinome in bladder cancer patients with the goal of identify therapeutic targets. Our analyses revealed amplification, overexpression, and elevated kinase activity of P21 (RAC1) activated kinase 4 (PAK4) in a subset of Bladder cancer (BLCA). Using bladder cancer cells, we confirmed the role of PAK4 in BLCA cell proliferation and invasion. Furthermore, we observed that a PAK4 inhibitor was effective in curtailing growth of BLCA cells. Transcriptomic analyses identified elevated expression of another kinase, protein tyrosine kinase 6 (PTK6), upon treatment with a PAK4 inhibitor and RNA interference of PAK4. Treatment with a combination of kinase inhibitors (vandetanib and dasatinib) showed enhanced sensitivity compared with either drug alone. Thus, PAK4 may be therapeutically actionable for a subset of MIBC patients with amplified and/or overexpressed PAK4 in their tumors. Our results also indicate that combined inhibition of PAK4 and PTK6 may overcome resistance to PAK4. These observations warrant clinical investigations with selected BLCA patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
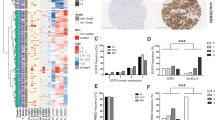
References
Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108.
von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, et al. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–8.
Grossman HB, Natale RB, Tangen CM, Speights VO, Vogelzang NJ, Trump DL, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N. Engl J Med. 2003;349:859–66.
Griffiths G, Hall R, Sylvester R, Raghavan D, Parmar MK. International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: long-term results of the BA06 30894 trial. J Clin Oncol. 2011;29:2171–7.
Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76.
Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N. Engl J Med. 2017;376:1015–26.
Sonpavde G. PD-1 and PD-L1 inhibitors as salvage therapy for urothelial carcinoma. N. Engl J Med. 2017;376:1073–4.
Balar AV, Castellano D, O’Donnell PH, Grivas P, Vuky J, Powles T, et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2017;18:1483–92.
Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65.
McConkey DJ, Choi W, Ochoa A, Siefker-Radtke A, Czerniak B, Dinney CP. Therapeutic opportunities in the intrinsic subtypes of muscle-invasive bladder cancer. Hematol Oncol Clin North Am. 2015;29:377–94. x-xi.
Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder. Cancer Cell 2017;171:540–56. e25.
Jacobs JJ, van Lohuizen M. Cellular memory of transcriptional states by polycomb-group proteins. Semin Cell Dev Biol. 1999;10:227–35.
Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol. 2001;2:409–21.
Sonpavde G, Jones BS, Bellmunt J, Choueiri TK, Sternberg CN. Future directions and targeted therapies in bladder cancer. Hematol Oncol Clin North Am. 2015;29:361–76. x.
Bellmunt J, Orsola A, Sonpavde G. Precision and predictive medicine in urothelial cancer: are we making progress? Eur Urol. 2015;68:547–49.
Poh A. Erdafitinib efficacious in bladder cancer. Cancer Discov. 2018;8:OF6.
Choudhury NJ, Campanile A, Antic T, Yap KL, Fitzpatrick CA, Wade JL, 3rd. et al. Afatinib activity in platinum-refractory metastatic urothelial carcinoma in patients with ERBB alterations. J Clin Oncol. 2016;34:2165–71.
Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122–a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–57.
Jian B, Li Z, Xiao D, He G, Bai L, Yang Q. Downregulation of microRNA-193-3p inhibits tumor proliferation migration and chemoresistance in human gastric cancer by regulating PTEN gene. Tumour Biol. 2016;37:8941–9.
Zhang HS, Zhang FJ, Li H, Liu Y, Du GY, Huang YH. Tanshinone A inhibits human esophageal cancer cell growth through miR-122-mediated PKM2 down-regulation. Arch Biochem Biophys. 2016;598:50–6.
Wang Y, Xing QF, Liu XQ, Guo ZJ, Li CY, Sun G. MiR-122 targets VEGFC in bladder cancer to inhibit tumor growth and angiogenesis. Am J Transl Res. 2016;8:3056–66.
Xu XL, Ye YL, Wu ZM, He QM, Tan L, Xiao KH, et al. Overexpression of PTK6 predicts poor prognosis in bladder cancer patients. J Cancer. 2017;8:3464–73.
Li N, Lopez MA, Linares M, Kumar S, Oliva S, Martinez-Lopez J. et al. Dual PAK4-NAMPT inhibition impacts growth and survival, and increases sensitivity to DNA-damaging agents in Waldenstrom Macroglobulinemia. Clin Cancer Res. 2018;25:369–77.
Phatak P, Burrows WM, Chesnick IE, Tulapurkar ME, Rao JN, Turner DJ, et al. MiR-199a-3p decreases esophageal cancer cell proliferation by targeting p21 activated kinase 4. Oncotarget. 2018;9:28391–407.
Mahlamaki EH, Kauraniemi P, Monni O, Wolf M, Hautaniemi S, Kallioniemi A. High-resolution genomic and expression profiling reveals 105 putative amplification target genes in pancreatic cancer. Neoplasia. 2004;6:432–9.
Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik JH, Ying H, et al. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci USA. 2008;105:19372–7.
Tyagi N, Bhardwaj A, Singh AP, McClellan S, Carter JE, Singh S. p-21 activated kinase 4 promotes proliferation and survival of pancreatic cancer cells through AKT- and ERK-dependent activation of NF-kappaB pathway. Oncotarget. 2014;5:8778–89.
Thillai K, Sarker D, Wells C. PAK4 pathway as a potential therapeutic target in pancreatic cancer. Future Oncol. 2018;14:579–82.
Yu W, Kanaan Y, Bae YK, Gabrielson E. Chromosomal changes in aggressive breast cancers with basal-like features. Cancer Genet Cytogenet. 2009;193:29–37.
Li SQ, Wang ZH, Mi XG, Liu L, Tan Y. MiR-199a/b-3p suppresses migration and invasion of breast cancer cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life. 2015;67:768–77.
Mao K, Lei D, Zhang H, You C. MicroRNA-485 inhibits malignant biological behaviour of glioblastoma cells by directly targeting PAK4. Int J Oncol. 2017;51:1521–32.
Zeng B, Shi W, Tan G. MiR-199a/b-3p inhibits gastric cancer cell proliferation via down-regulating PAK4/MEK/ERK signaling pathway. BMC Cancer. 2018;18:34.
Callegari E, D’Abundo L, Guerriero P, Simioni C, Elamin BK, Russo M, et al. miR-199a-3p Modulates MTOR and PAK4 Pathways and Inhibits Tumor Growth in a Hepatocellular Carcinoma Transgenic Mouse Model. Mol Ther Nucleic Acids. 2018;11:485–93.
He LF, Xu HW, Chen M, Xian ZR, Wen XF, Chen MN, et al. Activated-PAK4 predicts worse prognosis in breast cancer and promotes tumorigenesis through activation of PI3K/AKT signaling. Oncotarget. 2017;8:17573–85.
Xu HT, Lai WL, Liu HF, Wong LL, Ng IO, Ching YP. PAK4 phosphorylates p53 at serine 215 to promote liver cancer metastasis. Cancer Res. 2016;76:5732–42.
Park JJ, Park MH, Oh EH, Soung NK, Lee SJ, Jung JK. et al. The p21-activated kinase 4-Slug transcription factor axis promotes epithelial-mesenchymal transition and worsens prognosis in prostate cancer. Oncogene. 2018;37:5147–59.
Li Y, Zhang H, Zhao Y, Wang C, Cheng Z, Tang L. et al. A mandatory role of nuclear PAK4-LIFR axis in breast-to-bone metastasis of ERalpha-positive breast cancer cells. Oncogene. 2018;38:808–21.
Petrylak DP, Balar AV, O’Donnell PH, McGregor BA, Heath EI, Yu EY, et al. EV-201: results of enfortumab vedotin monotherapy for locally advanced or metastatic urothelial cancer previously treated with platinum and immune checkpoint inhibitors. J Clin Oncol. 2019;37:4505-.
Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl J Med. 2019;381:338–48.
Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl J Med. 2015;372:30–9.
Rogers MS, Foley MA, Crotty TB, Hartmann LC, Ingle JN, Roche PC, et al. Loss of immunoreactivity for human calmodulin-like protein is an early event in breast cancer development. Neoplasia. 1999;1:220–5.
Bennett RD, Pittelkow MR, Strehler EE. Immunolocalization of the tumor-sensitive calmodulin-like protein CALML3 in normal human skin and hyperproliferative skin disorders. PLoS ONE. 2013;8:e62347.
Brooks MD, Bennett RD, Weaver AL, Sebo TJ, Eckert SE, Strehler EE, et al. Human calmodulin-like protein CALML3: a novel marker for normal oral squamous mucosa that is downregulated in malignant transformation. Int J Dent. 2013;2013:592843.
Conrad C, Benzel J, Dorzweiler K, Cook L, Schlomann U, Zarbock A, et al. ADAM8 in invasive cancers: links to tumor progression, metastasis, and chemoresistance. Clin Sci. 2019;133:83–99.
Ravipaty S, Wu W, Dalvi A, Tanna N, Andreazi J, Friss T, et al. Clinical validation of a serum protein panel (FLNA, FLNB and KRT19) for diagnosis of prostate cancer. J Mol Biomark Diagn. 2017;8:323.
Iguchi Y, Ishihara S, Uchida Y, Tajima K, Mizutani T, Kawabata K, et al. Filamin B enhances the invasiveness of cancer cells into 3D Collagen Matrices. Cell Struct Funct. 2015;40:61–7.
Chen C, Shan H. Keratin 6A gene silencing suppresses cell invasion and metastasis of nasopharyngeal carcinoma via the betacatenin cascade. Mol Med Rep. 2019;19:3477–84.
Ricciardelli C, Lokman NA, Pyragius CE, Ween MP, Macpherson AM, Ruszkiewicz A, et al. Keratin 5 overexpression is associated with serous ovarian cancer recurrence and chemotherapy resistance. Oncotarget. 2017;8:17819–32.
Xiao J, Lu X, Chen X, Zou Y, Liu A, Li W, et al. Eight potential biomarkers for distinguishing between lung adenocarcinoma and squamous cell carcinoma. Oncotarget. 2017;8:71759–71.
Puvirajesinghe TM, Bertucci F, Jain A, Scerbo P, Belotti E, Audebert S, et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat Commun. 2016;7:10318.
Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, et al. Activating smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–2.
Huang XH, Chen JS, Wang Q, Chen XL, Wen L, Chen LZ, et al. miR-338-3p suppresses invasion of liver cancer cell by targeting smoothened. J Pathol. 2011;225:463–72.
Ye L, Li F, Song Y, Yu D, Xiong Z, Li Y, et al. Overexpression of CDCA7 predicts poor prognosis and induces EZH2-mediated progression of triple-negative breast cancer. Int J Cancer. 2018;143:2602–13.
Wang H, Ye L, Xing Z, Li H, Lv T, Liu H, et al. CDCA7 promotes lung adenocarcinoma proliferation via regulating the cell cycle. Pathol Res Pr. 2019;215:152559.
Grizzle WEBW, Fredenburgh J. Safety in biomedical and other laboratories. In: Patrinos G, Ansorg W, Editors. Molecular Diagnostics. USA: Elsevier Academic Press; Chapter 33, pp 421–8, 2005.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45.
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25.
Anderson JC, Willey CD, Mehta A, Welaya K, Chen D, Duarte CW, et al. High throughput kinomic profiling of human clear cell renal cell carcinoma identifies kinase activity dependent molecular subtypes. PLoS ONE. 2015;10:e0139267.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93.
Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Chakravarthi B, Rodriguez Pena MDC, Agarwal S, Chandrashekar DS, Hodigere Balasubramanya SA, Jabboure FJ, et al. A role for de novo purine metabolic enzyme PAICS in bladder cancer progression. Neoplasia. 2018;20:894–904.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11.
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–6.
Hao JQ, Li Q, Xu SP, Shen YX, Sun GY. Effect of lumiracoxib on proliferation and apoptosis of human nonsmall cell lung cancer cells in vitro. Chin Med J. 2008;121:602–7.
Acknowledgements
This study was supported in part by institutional funds (Department of Pathology and School of Medicine of the University of Alabama at Birmingham) awarded to SV. IBAB is supported by the Department of IT, BT and S&T, Government of Karnataka, India. The authors thank Dr Donald Hill for critical reading and editing of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
GS was a consultant for BMS, Exelixis, Bayer, Sanofi, Pfizer, Novartis, Eisai, Janssen, Amgen, AstraZeneca, Merck, Genentech, Astellas/Agensys; Research support to institution from Bayer, Amgen, Boehringer-Ingelheim, Merck, Sanofi, Pfizer; Author for Up-to-date; Speaker for Clinical Care Options, Physicians Education Resource (PER), Research to Practice (RTP), Onclive. CW was a consultant for Varian Medical Systems and LifeNet Health, Inc. AB and SD received financial support from Shodhaka LS Pvt. Ltd. KKA is the founder and director of Shodhaka LS Pvt. Ltd. GJN served as a consultant to Genentec. ESY was a consultant for Astrazeneca; Research support to institution from Astrazeneca, Eli Lilly, Novartis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chandrashekar, D.S., Chakravarthi, B.V.K., Robinson, A.D. et al. Therapeutically actionable PAK4 is amplified, overexpressed, and involved in bladder cancer progression. Oncogene 39, 4077–4091 (2020). https://doi.org/10.1038/s41388-020-1275-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1275-7
This article is cited by
-
Circ_0000235 targets MCT4 to promote glycolysis and progression of bladder cancer by sponging miR-330-5p
Cell Death Discovery (2023)
-
The role of p21-activated kinase 4 in the progression of oral squamous cell carcinoma by targeting PI3K–AKT signaling pathway
Clinical and Translational Oncology (2023)