Abstract
Context
Anthropogenic activities have detrimental impacts on natural habitats and the species inhabiting them. In particular, habitat fragmentation has a profound effect on the dynamics and structure of natural populations and the species’ probability of persistence.
Objectives
In this study, we examined which factors determine the population structure of Ctenomys species (tuco-tucos) at a local scale, evaluating the effects of natural and anthropic barriers on population divergence.
Methods
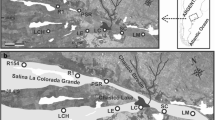
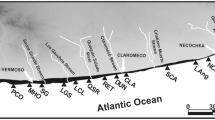
We sampled tuco-tucos at 28 localities and genotyped 231 individuals at 11 microsatellite loci. Additionally, we built six spatial layers that describe the landscape inhabited by tuco-tucos, to evaluate the effects of habitat traits in the movement of individuals. We applied Bayesian clustering methods to infer the population structure, and landscape genetic tools to understand how landscape traits affect this structure.
Results
We detected a high degree of population structure, even at a small spatial scale. Genetic structure seems to be influenced not only by current landscape configuration but also by their recent evolution. Altitude was the main contributing factor explaining this structure, with independent populations restricted to different sandy elevations in the region. However, anthropic activities were also shown to have had a significant effect on the differentiation among populations.
Conclusions
The accelerated transformation process that the region is undergoing strongly conditions the dynamics of population differentiation in Ctenomys and reduces prospects of viability for the species. Our findings underscore the importance of incorporating variables that describe the temporal component of habitat changes in landscapes experiencing intense and recent transformation processes.





Similar content being viewed by others
References
Akaike H (1974) A new look at the statistical model identification. Selected papers of Hirotugu Akaike. Springer, New York, pp 215–222
Andren H (1994) Effects of habitat fragmentation on birds and mammals in landscapes with different proportions of suitable habitat: a review. Oikos 71:355–366
Andren H, Delin A, Seiler A (1997) Population response to landscape changes depends on specialization to different landscape elements. Oikos 80:193–196
Antinuchi CD, Zenuto RR, Luna F, Cutrera AP, Perisinotti P, Busch C (2006) Energy budget in subterranean rodents: insights from the tuco-tuco Ctenomys talarum (Rodentia: Ctenomyidae). In: Kelt DA, Salazar-Bravo JA, Patton JL (eds) The quintessential naturalist: honoring the life and legacy of Oliver Pearson. University of California Press, Berkeley, pp 111–140
Augusto L, Ranger J, Binkley D, Rothe A (2002) Impact of several common tree species of European temperate forests on soil fertility. Ann For Sci 59:233–253
Bates D, Maechler M, Bolker B, and Walker S (2014) lme4: linear mixed-effects models using Eigen and S4. R package version 1.1-7
Beninde J, Feldmeier S, Werner M, Peroverde D, Schulte U, Hochkirch A, Veith M (2016) Cityscape genetics: structural vs. functional connectivity of an urban lizard population. Mol Ecol 25:4984–5000
Beninde J, Feldmeier S, Veith M, Hochkirch A (2018) Admixture of hybrid swarms of native and introduced lizards in cities is determined by the cityscape structure and invasion history. Proc R Soc B 285:20180143
Botana MI, Fernández SE (2018) Transformaciones territoriales en los Esteros del Iberá. In I Jornadas de Investigación “Ríos urbanos. Nuevas perspectivas para el estudio, diseño y gestión de los territorios fluviales". La Plata/Gral San Martín
Busch C, Antinuchi CD, del Valle JC, Kittlein MJ, Malizia AI, Vassallo AI, Zenuto RR (2000) Population ecology of subterranean rodents. In: Lacey EA, Patton JL, Cameron GN (eds) Life underground: the biology of subterranean rodents. University of Chicago Press, Chicago, pp 183–226
Caraballo DA, Abruzzese GA, Rossi MS (2012) Diversity of tuco-tucos (Ctenomys, Rodentia) in the Northeastern wetlands from Argentina: mitochondrial phylogeny and chromosomal evolution. Genetica 140:125–136
Clark-Laboratories (1999) Idrisi32. Clark University, Worcester
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: estimating gene flow with distance. J Agric Biol Environ Stat 7:361–372
Collinge SK (1996) Ecological consequences of habitat fragmentation: implications for landscape architecture and planning. Landsc Urban Plan 36:59–77
Crooks KR, Burdett CL, Theobald DM, King SRB, Di Marco M, Rondinini C, Boitani L (2017) Quantification of habitat fragmentation reveals extinction risk in terrestrial mammals. Proc Natl Acad Sci 114:7635–7640
Cutrera AP, Lacey EA, Busch C (2005) Genetic structure in a solitary rodent (Ctenomys talarum): implications for kinship and dispersal. Mol Ecol 14:2511–2523
De Freitas TRO (2016) Family ctenomyidae (Tuco-tucos). In: Wilson DE, Lacher TE, Mittermeier RA (eds) The handbook of mammals of the world. Lagomorphs and Rodents I Lynx Edicions, Barcelona, pp 498–534
Dudaniec RY, Rhodes JR, Worthington Wilmer J, Lyons M, Lee KE, McAlpine CA, Carrick FN (2013) Using multilevel models to identify drivers of landscape-genetic structure among management areas. Mol Ecol 22:3752–3765
Earl DA, von Holdt B (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Epps CW, Keyghobadi N (2015) Landscape genetics in a changing world: disentangling historical and contemporary influences and inferring change. Mol Ecol 24:6021–6040
Esperandio IB, Ascensão F, Kindel A, Tchaicka L, de Freitas TRO (2019) Do roads act as a barrier to gene flow of subterranean small mammals? A case study with Ctenomys minutus. Conserv Genet 20:385–393
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewers RM, Didham RK (2006) Confounding factors in the detection of species responses to habitat fragmentation. Biol Rev 81:117–142
Excoffier L, Laval G, Schneider S (2005) ARLEQUIN ver. 3.0: an integrated software packaged for population genetics data analysis. Evol Bioinform 1:47–50
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fernández-Stolz GP, Stolz JFB, De Freitas TRO (2007) Bottlenecks and dispersal in the tuco-tuco das dunas, Ctenomys flamarioni (Rodentia: Ctenomyidae), in southern Brazil. J Mammal 88:935–945
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618
Gallardo LI, Coronel JM, Poi ASG (2019) Urban rain-fed lakes: macro-invertebrate assemblages associated with Egeria najas as indicators of biological integrity in wetlands of Corrientes Province (Argentina). Biodivers Conserv 29:1–20
García L, Ponsà M, Egozcue J, García M (2000) Cytogenetic variation in Ctenomys perrensi (Rodentia, Octodontidae). Biol J Lin Soc 71:615–624
Giménez MD, Mirol PM, Bidau CJ, Searle JB (2002) Molecular analysis of populations of Ctenomys (Caviomorpha, Rodentia) with high karyotypic variability. Cytogenet Genome Res 96:130–136
Gómez Fernández MJ, Boston ES, Gaggiotti OE, Kittlein MJ, Mirol PM (2016) Influence of environmental heterogeneity on the distribution and persistence of a subterranean rodent in a highly unstable landscape. Genetica 144:711–722
Gómez Fernández MJ, Gaggiotti OE, Mirol PM (2012) The evolution of a highly speciose group in a changing environment: are we witnessing speciation in the Iberá wetlands? Mol Ecol 21:3266–3282
Gonçalves GL, De Freitas TRO (2009) Intraspecific variation and genetic differentiation of the collared tuco-tuco (Ctenomys torquatus) in southern Brazil. J Mammal 90:1020–1031
Guillot G, Mortier F, Estoup A (2005) GENELAND: a computer package for landscape genetics. Mol Ecol Notes 5:712–715
Guillot G, Renaud S, Ledevin R, Michaux J, Claude J (2012) A unifying model for the analysis of phenotypic, genetic, and geographic data. Syst Biol 61:897–911
Guo S, Thompson E (1992) Performing the exact test of Hardy-Weinberg proportion for multiples alleles. Biometrics 48:361–372
Haddad NM, Brudvig LA, Clobert J, Davies KF, Gonzalez A, Holt RD, Townshend JR (2015) Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci Adv 1:1–9
Hanski I (1999) Metapopulation ecology. Oxford University Press, Oxford
Hantak MM, Page RB, Converse PE, Anthony CD, Hickerson CAM, and Kuchta SR (2019). Do genetic structure and landscape heterogeneity impact color morph frequency in a polymorphic salamander?. Ecography.
Holzhauer SIJ, Ekschmitt K, Sander AC, Dauber J, Wolters V (2006) Effect of historic landscape change on the genetic structure of the bush-cricket Metrioptera roeseli. Landsc Ecol 21:891–899
Janes JK, Miller JM, Dupuis JR, Malenfant RM, Gorrell JC, Cullingham CI, Andrew RL (2017) The K= 2 conundrum. Mol Ecol 26:3594–3602
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Lacey EA (2000) Spatial and social systems of subterranean rodents. In: Lacey EA, Patton JL, Cameron GN (eds) Life underground: the biology of subterranean rodents. University of Chicago Press, Chicago, pp 257–296
Lacey EA (2001) Microsatellite variation in solitary and social tuco tucos: molecular properties and population dynamics. Heredity 86:628–637
Lacey EA, Maldonado JE, Clabaugh JP, Matocq MD (1999) Interspecific variation in microsatellites isolated from tuco-tucos (Rodentia: Ctenomyidae). Mol Ecol 8:1753–1768
Landguth EL, Cushman SA, Schwartz MK, McKelvey KS, Murphy M, Luikart G (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191
Lanzone C, Giménez MD, Santos JL, Bidau CJ (2007) Meiotic effects of Robertsonian translocations in tuco-tucos of the Ctenomys perrensi superspecies (Rodentia: Ctenomyidae). Caryologia 60:233–244
Laurance WF, Bierregaard RO (1997) Tropical forest remnants: ecology, management, and conservation of fragmented communities. University of Chicago Press, Chicago
Lillesand T, Kiefer RW, Chipman J (2014) Remote sensing and image interpretation. Wiley, New York
Mantel N (1967) The detection of disease clustering and a generalized regression approaches. Can Res 27:209–220
Mapelli FJ, Kittlein MJ (2009) Influence of patch and landscape characteristics on the distribution of the subterranean rodent Ctenomys porteousi. Landsc Ecol 24:723–733
Mapelli FJ, Mora MS, Mirol PM, Kittlein MJ (2012a) Population structure and landscape genetics in the endangered subterranean rodent Ctenomys porteousi. Conserv Genet 13:165–181
Mapelli FJ, Mora MS, Mirol PM, Kittlein MJ (2012b) Effects of Quaternary climatic changes on the phylogeography and historical demography of the subterranean rodent Ctenomys porteousi. J Zool 286:48–57
Milanesi P, Holderegger R, Caniglia R, Fabbri E, Randi E (2016) Different habitat suitability models yield different least-cost path distances for landscape genetic analysis. Basic Appl Ecol 17:61–71
Miller MP (2005) Alleles In Space (AIS): computer software for the joint analysis of interindividual spatial and genetic information. J Hered 96:722–724
Miller SA, Dikes DD, Polesky HH (1988) A simple salting procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:215
Mirol P, Gimenez MD, Searle JB, Bidau CJ, Faulkes CG (2010) Population and species boundaries in the South American subterranean rodent Ctenomys in a dynamic environment. Biol J Lin Soc 100:368–383
Mora MS, Lessa EP, Cutrera AP, Kittlein MJ, Vassallo AI (2007) Phylogeographical structure in the subterranean tuco-tuco Ctenomys talarum (Rodentia: Ctenomyidae): contrasting the demographic consequences of regional and habitat-specific histories. Mol Ecol 16:3453–3465
Mora MS, Lessa EP, Kittlein MJ, Vassallo AI (2006) Phylogeography of the subterranean rodent Ctenomys australis in sand-dune habitats: evidence of population expansion. J Mammal 87:1192–1203
Mora MS, Mapelli FJ, Gaggiotti OE, Kittlein MJ, Lessa EP (2010) Dispersal and population structure at different spatial scales in the subterranean rodent Ctenomys australis. BMC Genet 11:9
Mora MS, Mapelli FJ, López A, Gómez Fernández MJ, Mirol PM, Kittlein MJ (2017) Landscape genetics in the subterranean rodent Ctenomys “chasiquensis” associated with highly disturbed habitats from the southeastern Pampas region, Argentina. Genetica 145:575–591
Muscarella R, Galante PJ, Soley-Guardia M, Boria RA, Kass JM, Uriarte M, Anderson RP (2014) ENM eval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol Evol 5:1198–1205
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Ortells MO, Contreras JR, Reig OA (1990) New Ctenomys karyotypes (Rodentia, Octodontidae) from north-eastern Argentina and from Paraguay confirm the extreme chromosomal multiformity of the genus. Genetica 82:189–201
Pannell JR, Charlesworth B (1999) Neutral genetic diversity in a metapopulation with recurrent local extinction and recolonization. Evolution 53:664–676
Smouse RPP, Peakall R (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 28:2537–2539
Peterman WE (2018) ResistanceGA: an R package for the optimization of resistance surfaces using genetic algorithms. BioRxiv. https://doi.org/10.1101/007575
Peterman WE, Connette GM, Semlitsch RD, Eggert LS (2014) Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol Ecol 23:2402–2413
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pritchard JK, Wen W (2003) Documentation for STRUCTURE software: version 2. https://pritch.bsd.uchicago.edu
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Radosavljevic A, Anderson RP (2014) Making better Maxent models of species distributions: complexity, overfitting and evaluation. J Biogeogr 41:629–643
Richmond JQ, Wood DA, Westphal MF, Vandergast AG, Leaché AD, Saslaw LR, Butterfield HS, Fisher RN (2017) Persistence of historical population structure in an endangered species despite near-complete biome conversion in California's San Joaquin Desert. Mol Ecol 26:3618–3635
Rice WR (1989) Analysing tables of statistical tests. Evolution 43:223–225
Rouse JW, Haas RH, Schell JA, Deering DW, Harlan JC (1974) Monitoring the vernal advancement of retrogradation of natural vegetation. Technical report, NASA/ GSFC, Type III, Final Report, Greenbelt, MD, USA
Rousset F (2008) Genepop: a complete re-implementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Safner T, Miller MP, McRae BH, Fortin MJ, Manel S (2011) Comparison of Bayesian clustering and edge detection methods for inferring boundaries in landscape genetics. Int J Mol Sci 12:865–889
Scrucca L (2013) GA: A package for genetic algorithms in R. J Stat Softw 53:1–37
Sikes RS, Animal Care and Use Committee of the American Society of Mammalogists (2016) 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal 97:663–688
Spear SF, Storfer A (2008) Landscape genetic structure of coastal tailed frogs (Ascaphus truei) in protected vs. managed forests. Mol Ecol 17:4642–4656
Spear SF, Balkenhol N, Fortin MJ, McRae BH, Scribner KIM (2010) Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Mol Ecol 19:3576–3591
Tallmon DA, Bellemain E, Swenson J, Taberlet P (2004) Genetic monitoring of Scandinavian brown bear effective population size and immigration. J Wildl Manag 68:960–965
Vallejos VH, Schnake VP (2014. Colonia Santa Rosa: transformaciones territoriales y rol del Estado. Departamento de Concepción, Provincia de Corrientes. X Jornadas de Investigación del Departamento de Geografía. Facultad de Ciencias Humanas. Universidad Nacional de Río Cuarto
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Varvio S, Chakraborty R, Nei M (1986) Genetic variation in subdivided populations and conservation genetics. Heredity 57:189–198
Wang JL (2004) Application of the one-migrant-per-generation rule to conservation and management. Conserv Biol 18:332–343
Wang YH, Yang KC, Bridgman CL, Lin LK (2008) Habitat suitability modelling to correlate gene flow with landscape connectivity. Landsc Ecol 23:989–1000
Watling JI, Donnelly MA (2006) Fragments as islands: a synthesis of faunal responses to habitat patchiness. Conserv Biol 20:1016–1025
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock M (1992) Temporal fluctuations in demographic parameters and the genetic variance among populations. Evolution 46:608–615
Wilson MC, Chen XY, Corlett RT, Didham RK, DingHolt PRD, Yu M (2016) Habitat fragmentation and biodiversity conservation: key findings and future challenges. Landsc Ecol 31:219–227
Wlasiuk G, Garza JC, Lessa EP (2003) Genetic and geographic differentiation in the Rio Negro tuco-tuco (Ctenomys rionegrensis): inferring the roles, of migration and drift from multiple genetic markers. Evolution 57:913–926
Zeng DH, Hu YL, Chang SX, Fan ZP (2009) Land cover change effects on soil chemical and biological properties after planting Mongolian pine (Pinus sylvestris var. mongolica) in sandy lands in Keerqin, northeastern China. Plant Soil 317:121–133
Acknowledgements
We wish to extend our gratitude to Matias S. Mora and Yanina Perez, who collaborated during field work. We thank the members of GECoBi for their valuable input and interpretation of results. Finallly we want to thank the citizens of San Miguel, Paraje Caimán, Colonia Montaña, Santa Bárbara, Silvero-Cué, Yataití-Poí and Colonia Capilla for their hospitality and warm predisposition. Financial support was provided by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP-0138) and FONCYT (PICT-1551).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mapelli, F.J., Boston, E.S.M., Fameli, A. et al. Fragmenting fragments: landscape genetics of a subterranean rodent (Mammalia, Ctenomyidae) living in a human-impacted wetland. Landscape Ecol 35, 1089–1106 (2020). https://doi.org/10.1007/s10980-020-01001-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-020-01001-z